- Title
-
Bioinformatic analysis and functional predictions of selected regeneration-associated transcripts expressed by zebrafish microglia
- Authors
- Issaka Salia, O., Mitchell, D.M.
- Source
- Full text @ BMC Genomics
|
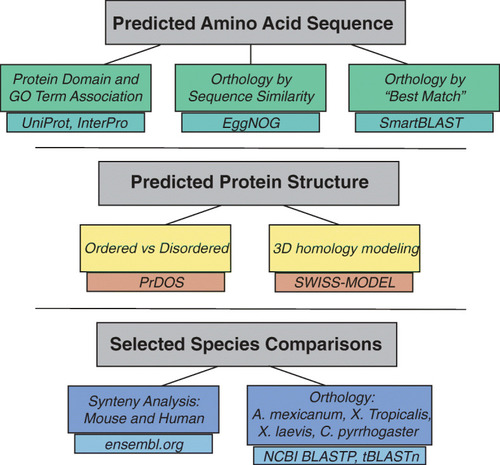
Overview of Bioinformatic Analysis for Functional Predictions. The diagram shows an overview of the bioinformatic analyses performed in order to make functional predictions about the genes of interest based on ( |
|
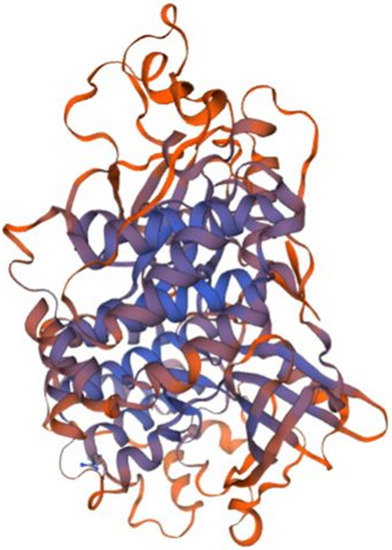
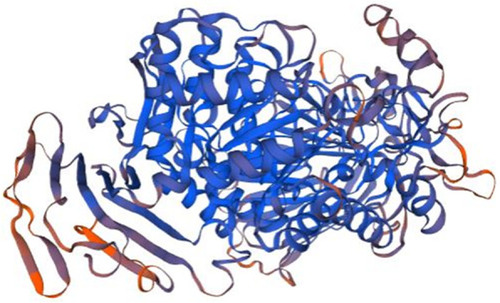
Homology model of P1 putative kinase domain. The kinase domain of Receptor-interacting serine/threonine-protein kinase 2 (RIPK2, 6fu5.1.B in the rcsb protein database) is the template used for the homology modelling of P1. The X-RAY diffraction 3.26 Å was used to determine the experimental structure of 6fu5.1 [ |
|
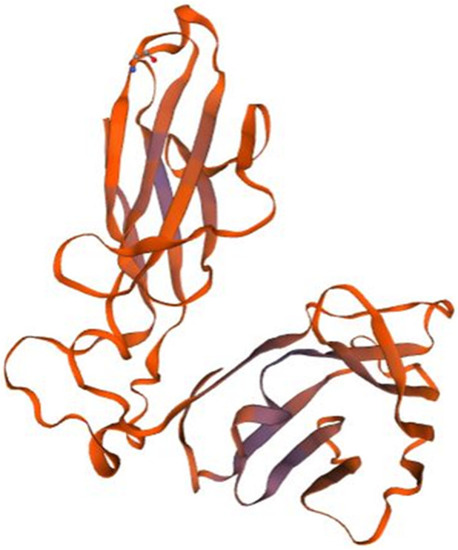
Homology model of P3. T cell receptor beta chain (3of6.1.A in the rcsb protein database) is the template used for the homology modelling of P3. The homology model starts from the P3 residue N°32 (THR, Threonine) and ends with the residue N° 245 (THR, Threonine). The X-RAY diffraction 2.80 Å was used to determine the experimental structure of 3of6.1.A [ |
|
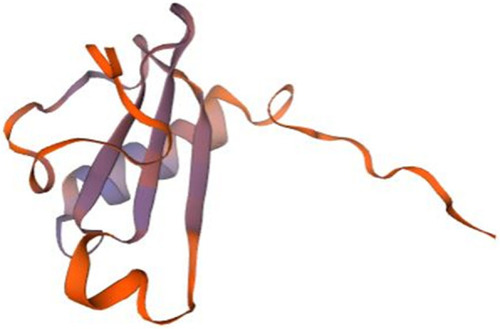
Homology model of P10 chemokine interleukin-8-like domain. Lymphotactin (1j8i.1.A in the rcsb protein database) is the template used for the homology modelling of P10. The homology model starts from P10 residue N°24 (GLU, Glutamic acid) and ends with the residue N° 102 (SER, Serine). The NMR spectroscopy was used to determine the experimental structure of 1j8i.1.A [ |
|
Homology model of P11. Maltase-glucoamylase, intestinal (3top.1.A in the rcsb protein database) is the template used for the homology modelling of P11. The X-RAY diffraction 2.9 Å was used to determine the experimental structure of 3top.1.A [ |
|
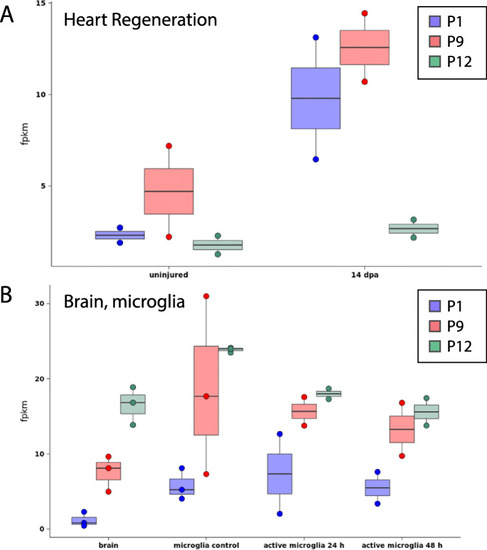
Expression level of selected zebrafish genes in other published studies. Expression level of selected zebrafish genes (P1, P9, and P12) in other published RNA-seq datasets of ( |