- Title
-
A zebrafish functional genomics model to investigate the role of human A20 variants in vivo
- Authors
- Cultrone, D., Zammit, N.W., Self, E., Postert, B., Han, J.Z.R., Bailey, J., Warren, J., Croucher, D.R., Kikuchi, K., Bogdanovic, O., Chtanova, T., Hesselson, D., Grey, S.T.
- Source
- Full text @ Sci. Rep.
|
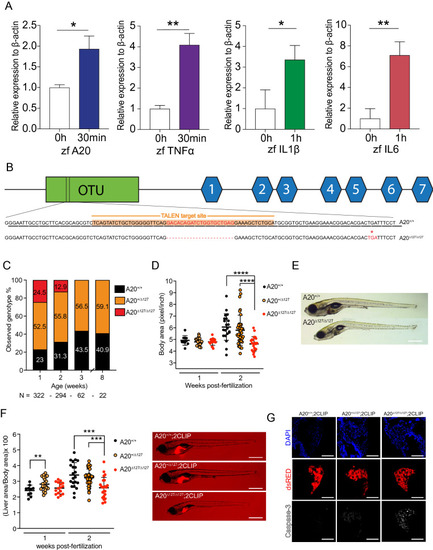
A20-deficient zebrafish fail to thrive. ( EXPRESSION / LABELING:
PHENOTYPE:
|
|
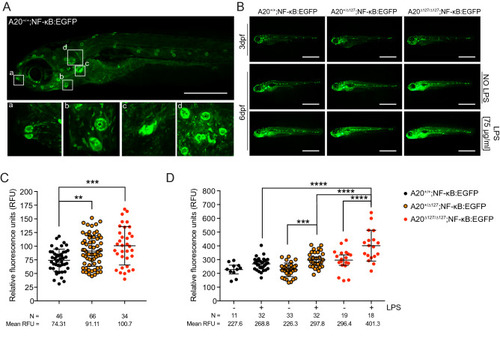
A20 is necessary to maintain inflammatory homeostasis in zebrafish. ( EXPRESSION / LABELING:
PHENOTYPE:
|
|
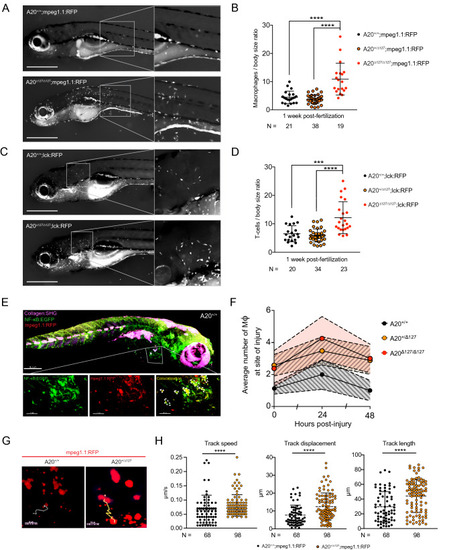
Impact of A20 deletion on macrophage activity in zebrafish. ( PHENOTYPE:
|
|
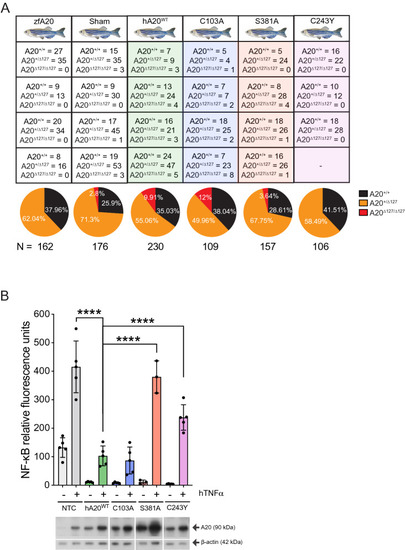
The impact of human A20 mutants on A20∆127/∆127 zebrafish survival. ( |