- Title
-
Gsx2 is required for specification of neurons in the inferior olivary nuclei from Ptf1a-expressing neural progenitors in zebrafish
- Authors
- Itoh, T., Takeuchi, M., Sakagami, M., Asakawa, K., Sumiyama, K., Kawakami, K., Shimizu, T., Hibi, M.
- Source
- Full text @ Development
|
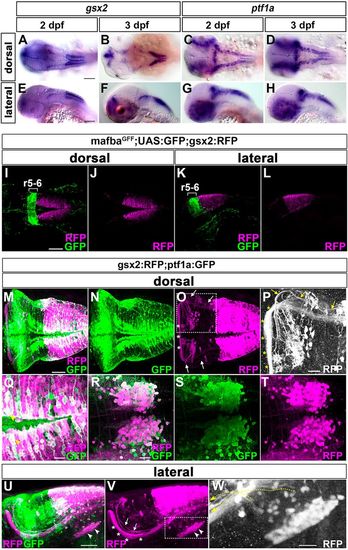
gsx2 and ptf1a are co-expressed in IO progenitors. (A,B,E,F) Expression of gsx2 in 2 and 3 dpf larvae. (C,D,G,H) Expression of ptf1a in 2 and 3 dpf larvae. (I-L) 3 dpf mafbaGFF;UAS:GFP;gsx2:RFP larvae (n=2) were immunostained using anti-DsRed (RFP, magenta) and anti-GFP (green) antibodies. r5-6, rhombomeres 5 and 6. (M-W) 5 dpf gsx2:RFP;ptf1a:GFP larvae were immunostained using anti-DsRed (magenta) and anti-GFP (green) antibodies. Dorsal views (M-T) and lateral views (U-W). (P) Higher magnification of the dotted box in O. (Q) Higher magnification of the dotted box in M (dorsal optical section). (R-T) Expression of GFP and/or RFP in the IO region. (W) Higher magnification of the dotted box in V. White and yellow arrows, and white arrowheads indicate CFs and IO neurons, respectively. Yellow arrowheads indicate both GFP+ and RFP+ cells. Asterisks indicate gsx2:RFP+ axons that are potentially axons from the nucleus commissure of Wallenberg, which sends mossy fibers projecting to GCs. Dotted yellow arrows in W indicate axons from IO neurons. Scale bars: 100 µm in A,E (apply to A-H) and I (applies to I-L); 50 µm in M (applies to M-O) and U (applies to U,V); 20 µm in P-R (bar in R applies to R-T) and W. EXPRESSION / LABELING:
|
|
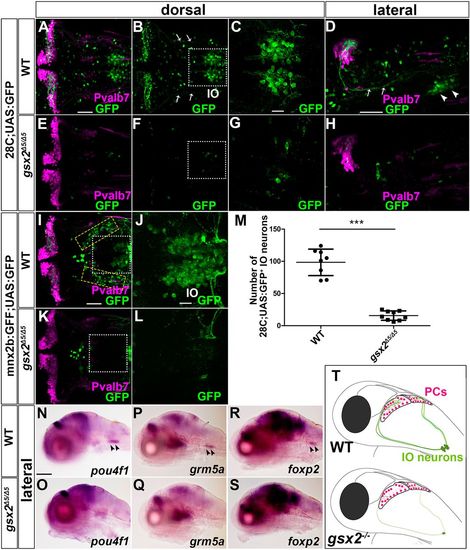
Strong reduction or loss of IO neurons in gsx2 mutants. (A-H) 5 dpf wild-type (A-D, n=10) and gsx2Δ5/Δ5 (E-H, n=10) hspGFFDMC28C;UAS:GFP (28C;UAS:GFP) larvae immunostained using anti-Pvalb7 (magenta) and anti-GFP (green) antibodies. Arrows and arrowheads indicate CFs and IO neurons, respectively. (C,G) Higher magnification of the boxes in B and F. (I-L) 5 dpf wild-type (I,J, n=6) and gsx2Δ5/Δ5 (K,L, n=7) mnx2b:GFF;UAS:GFP larvae. GFP+ cells in the yellow dotted boxes were fewer or absent in gsx2Δ5/Δ5 larvae. (J,L) Higher magnification of the boxes in I and K. (M) Number of 28C;UAS:GFP+ IO neurons in wild-type and gsx2Δ5/Δ5 larvae. 28C;UAS:GFP+ IO neurons were significantly fewer in gsx2 mutant larvae compared with control larvae. ***P<0.001 (Student's t-test). Data are mean±s.e.m. with individual values indicated. (N-S) Expression of endogenous IO neuronal markers in wild type and gsx2Δ5/Δ5. (N,O) Expression of pou4f1 in wild type (n=4) and gsx2Δ5/Δ5 (n=4). (P,Q) Expression of grm5a in wild type (n=3) and gsx2Δ5/Δ5 (n=2). (R,S) Expression of foxp2 in wild type (n=5) and gsx2Δ5/Δ5 (n=5). (T) Illustration of PCs and IO neurons in wild-type and gsx2 mutant larvae. Scale bars: 100 µm in N (applies to N-S); 50 µm in A (applies to A,B,E,F), D (applies to D,H) and I (applies to I,K); 20 µm in C (applies to C,G) and J (applies to J,L). |
|
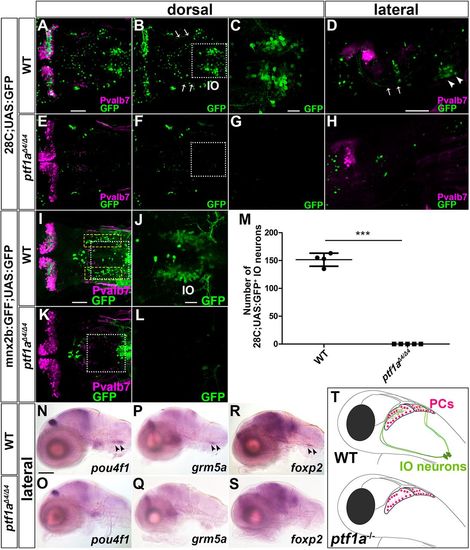
Loss of IO neurons in ptf1a mutants. (A-H) 5 dpf wild-type (A-D, n=5) and ptf1aΔ4/Δ4 (E-H, n=5) 28C;UAS:GFP larvae were immunostained using anti-Pvalb7 (magenta) and anti-GFP (green) antibodies. Arrows and arrowheads indicate CFs and IO neurons, respectively. (C,G) Higher magnification of the boxes in B and F. (I-L) 5 dpf wild-type (I-J n=2) and ptf1aΔ4/Δ4 (K-L n=4) mnx2b:GFF;UAS:GFP larvae. GFP+ cells in the yellow dotted boxes were fewer or absent in ptf1aΔ4/Δ4 larvae. (J,L) Higher magnification of the boxes in I and K. (M) Number of 28C;UAS:GFP+ IO neurons in wild-type and ptf1aΔ4/Δ4 larvae. 28C;UAS:GFP+ IO neurons were significantly reduced in ptf1a mutant larvae compared with control larvae. ***P<0.001 (Student's t-test). Data are mean± s.e.m. with individual values indicated. (N,O) Expression of pou4f1 in 5 dpf wild-type (n=4) and ptf1aΔ4/Δ4 (n=4) larvae. (P,Q) Expression of grm5a in 5 dpf wild-type (n=4) and ptf1aΔ4/Δ4 (n=3) larvae. (R,S) Expression of foxp2 in 5 dpf wild-type (n=5) and ptf1aΔ4/Δ4 (n=5) larvae. (T) Illustration of Purkinje cells and IO neurons in wild-type and ptf1a mutant larvae. Scale bars: 100 µm in N (applies to N-S); 50 µm in A (applies to A,B,E,F), D (applies to D,H) and I (applies to I,K); 20 µm in C (applies to C,G) and J (applies to J,L). |
|
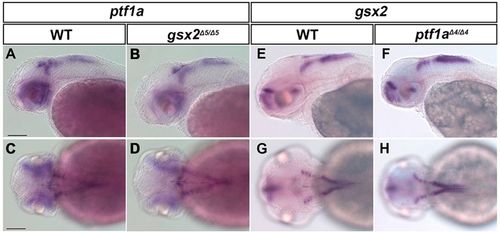
Expression of ptf1a and gsx2 was not affected in gsx2 and ptf1a mutants. (A-D) Expression of ptf1a in 2-dpf wild-type (A,C, n=2) and gsx2 mutant (B,D, n=4) larvae. (E-H) Expression of gsx2 in 2 dpf wild-type (E,G, n=2) and ptf1a mutant (F,H, n=3) larvae. Scale bars: 100 µm in A (applies to A,B,E,F) and C (applies to C,D,G,H). EXPRESSION / LABELING:
|
|
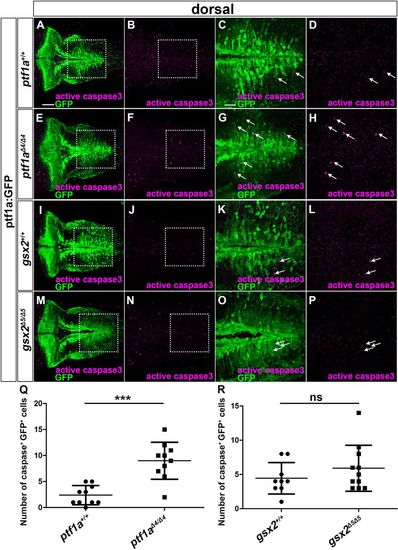
Apoptosis occurs in ptf1a but not gsx2 mutants. (A-P) 5 dpf ptf1a+/+ (A-D, n=10), ptf1aΔ4/Δ4 (E-H, n=10), gsx2+/+ (I-L, n=9) and gsx2Δ5/Δ5 (M-P, n=11) ptf1a:GFP larvae were stained using anti-GFP (green) and anti-cleaved caspase 3 (magenta) antibodies. (C,G,K,O) Higher magnification of the boxes in A,E,I,M. (D,H,L,P) Higher magnification of the boxes in B,F,J,N. Arrows indicate both GFP+ and cleaved caspase 3+ cells. (Q,R) Number of GFP+ and cleaved caspase 3+ cells in the caudal hindbrain in wild type, and the ptf1a and gsx2 mutants. ns, not significant; ***P<0.001 (Student's t-test for Q,R). Data are mean±s.e.m. with individual values indicated. Scale bars: 50 µm in A (applies to A,B,E,F,I,J,M,N); 20 µm in C (applies to C,D,G,H,K,L,O,P). PHENOTYPE:
|
|
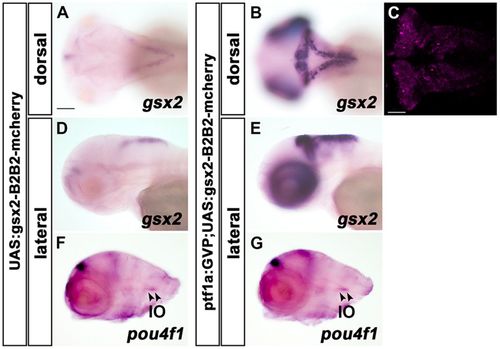
Ectopic gsx2 expression alone does not induce IO neurons. (A,B,D,E) Expression of gsx2 in 3 dpf Tg(UAS:gsx2BLRP-P2A-BirA-P2A-mCherry) (control, A,D, n=5) and TgBAC(ptf1a:Gal4-VP16);Tg(UAS:gsx2BLRP-P2A-BirA-P2A-mCherry) (B,E, n=5) larvae. The Tg(UAS:gsx2BLRP-P2A-BirA-P2A-mCherry) line harbors a transgene containing the open reading frame (ORF) of gsx2, biotin ligase recognition peptide (BLRP), 2A peptide sequences of porcine teschovirus-1 (P2A), biotin ligase (BirA) and mCherry genes. Expression of Gsx2 and mCherry can be driven by Gal4-VP16. (C) Expression of mCherry was induced where ptf1a was expressed in 3 dpf Tg larvae. (F,G) Expression of pou4f1 in 3 dpf control (F, n=3) and Tg(ptf1a:Gal4-VP16);Tg(UAS:gsx2BLRP-P2A-BirA-P2A-mCherry) (G, n=3) larvae. Although gsx2 was ectopically expressed in all ptf1a-expressing cells, it did not induce ectopic expression of pou4f1. Scale bars: 100 µm in A (applies to A,B,D-G); 50 µm in C. PHENOTYPE:
|
|
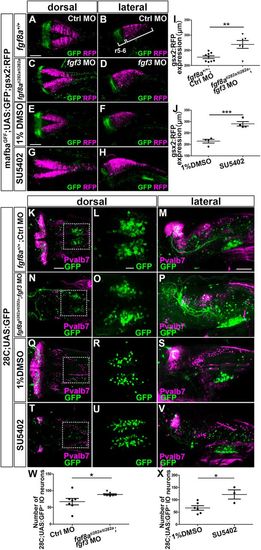
Fgf signal suppresses gsx2 expression and IO neuronal development. (A-D) 3 dpf control (fgf8a+/+) and fgf8ati282a/ti282a mafbaGFF;UAS:GFP;gsx2:RFP larvae that received an injection of control (Ctrl) MO and fgf3 MO, respectively, were stained using anti-RFP (magenta) and anti-GFP (green) antibodies. (E-H) mafbaGFF;UAS:GFP;gsx2:RFP were treated with 1% DMSO (E,F, n=4) or 200 µM SU5402 (G,H, n=4) from 6 to 22 hpf, fixed at 3 dpf, and stained using anti-RFP (magenta) and anti-GFP (green) antibodies. (I,J) Length of the gsx2:RFP+ hindbrain region (gsx2 expression) in fgf8a+/+; Ctrl MO (n=9) and fgf8ati282a/ti282a; fgf3MO (n=6) larvae (I), and in DMSO- (n=4) or 200 μM SU5402-treated (n=4) larvae (J). (K-P) Expression of GFP (green) and Pvalb7 (magenta) in control (fgf8a+/+) and fgf8ati282a/ti282a 28C;UAS:GFP larvae that received an injection of control MO and fgf3 MO (fgf3MO), respectively. (Q-V) 28C;UAS:GFP were treated with 1% DMSO (Q-S, n=6) or 5.0 µM SU5402 (T-V, n=3) from 6 to 22 hpf, fixed at 3 dpf, and stained using anti-Pvalb7 (magenta) and anti-GFP (green) antibodies. (L,O,R,U) Higher magnification of the boxes in K,N,Q,T. (W) Number of 28C;UAS:GFP+ IO neurons in fgf8a+/+;Ctrl MO (n=6) and fgf8ati282a/ti282a; fgf3 MO (n=5) larvae. (X) Number of 28C;UAS:GFP+ IO neurons in DMSO- (n=6) or 5.0 μM SU5402-treated (n=3) larvae. r5-6, rhombomeres 5 and 6. *P<0.05; **P<0.01; ***P<0.001 (Student's t-test for J and X, Welch's t-test for I and W). Data are mean±s.e.m. with individual values indicated. Scale bars: 100 µm in A (applies to A-D) and E (applies to E-H); 50 µm in K (applies to K,N,Q,T) and M (applies to M,P,S,V); 20 µm in L (applies to L,O,R,U). PHENOTYPE:
|
|
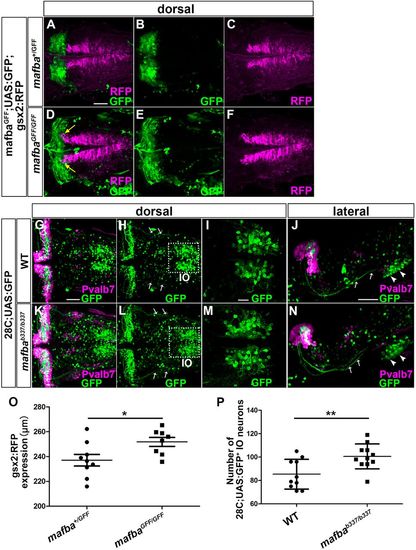
Mafba negatively controls gsx2 expression and IO neuronal development. (A-F) 3-dpf mafba+/GFF (A-C, n=9) and mafbaGFF/GFF (D-F, n=8);UAS:GFP;gsx2:RFP larvae were stained with anti-RFP (magenta) and anti-GFP (green) antibodies. Yellow arrows indicate that gsx2 expression extends to rhombomeres 5 and 6. (G-N) 5-dpf wild-type (G-J, n=10) and mafbab337/b337 28C;UAS:GFP (K-N, n=11) larvae were stained with anti-GFP (green) and anti-Pvalb7 (magenta) antibodies. Arrows and arrowheads indicate CFs and IO neurons, respectively. (O) Length of gsx2:RFP+ hindbrain region in mafba+/GFF and mafbaGFF/GFF. (P) Number of 28C;UAS:GFP+ IO neurons in wild-type and mafbab337/b337 larvae. *P<0.05; **P<0.01 (Student's t-test). Data are mean± s.e.m. with individual values indicated. Scale bars: 50 µm in A (applies A to F) and G (applies to G,H,K,L) and J (applies to J,N); 20 µm in I (applies to I,M). PHENOTYPE:
|
|
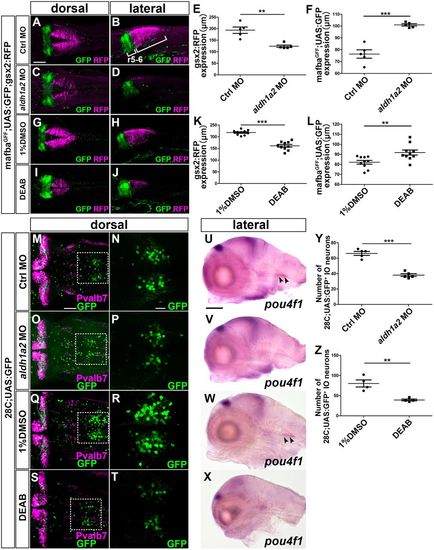
RA signal positively controls gsx2 expression and IO neuronal development. (A-D) 3 dpf control morphant (A,B, n=5) and aldh1a2 morphant (C,D, n=5) mafbaGFF;UAS;GFP;gsx2:RFP larvae were stained using anti-RFP (magenta) and anti-GFP (green) antibodies. (E,F) Extent of gsx2:RFP (E) and mafbaGFF;UAS:GFP expression (F) in the hindbrain of the control and aldh1a2 morphants. (G-J) mafbaGFF;UAS;GFP;gsx2:RFP larvae were treated with 1% DMSO (G,H, n=10) and 0.25 µM DEAB (I,J,H,I, n=10) from 6 to 22 hpf, fixed at 3 dpf, and stained using anti-GFP and anti-RFP antibodies. (K,L) Extent of gsx2:RFP (K) and mafbaGFF;UAS:GFP (L) expression in the hindbrain of larvae treated with DMSO or DEAB. (M-T) 5 dpf control morphant (M,N, n=5) and aldh1a2 morphant (O,P, n=5), DMSO- (Q,R, n=4) or DEAB-treated (S,T, n=4) 28C;UAS:GFP larvae were stained using anti-Pvalb7 (magenta) and anti-GFP (green) antibodies. (N,P,R,T) Higher magnification of the boxes in M,O,Q,S. (U-X) Expression of pou4f1 in 5 dpf control morphant (n=5), aldh1a2 morphant (n=5), DMSO-treated (n=3) or DEAB-treated (n=3) larvae. (Y) The number of 28C;UAS:GFP+ IO neurons in 5 dpf control morphant and aldh1a2 morphant larvae. (Z) Number of 28C;UAS:GFP+ IO neurons in 5 dpf larvae treated with DMSO and DEAB. **P<0.01; ***P<0.001 (Student's t-test). Data are mean±s.e.m. with individual values indicated. Scale bars: 100 µm in A (applies to A-D,G-J) and U (applies to U-X); 50 µm in M (applies to M,O,Q,S); 20 µm in N (applies to N,P,R,T). EXPRESSION / LABELING:
PHENOTYPE:
|
|
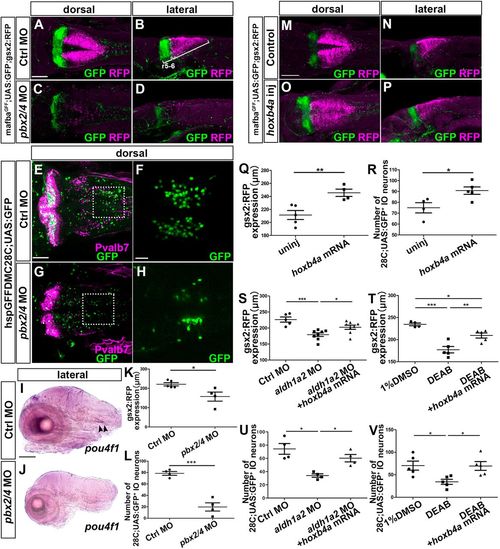
Hox genes are involved in gsx2 expression and IO neuronal development. (A-D) 3 dpf control morphant (A,B, n=5) and pbx2/4 morphant (C,D, n=4) mafbaGFF;UAS;GFP;gsx2:RFP larvae were stained using anti-RFP (magenta) and anti-GFP (green) antibodies. (E-H) 5 dpf control morphant (E,F, n=4) and pbx2/4 morphant (G,H, n=4) 28C;UAS:GFP larvae stained using anti-Pvalb7 (magenta) and anti-GFP (green) antibodies. (I,J) Expression of pou4f1 in 5 dpf control morphant (I) and pbx2/4 morphant (J) larvae. The pou4f1 signal is marked by arrowheads. (K,L) Length of the gsx2:RFP+ hindbrain region (K) and number of 28C;UAS:GFP+ IO neurons (L) in control and pbx2/4 morphant larvae. (M-P) 3 dpf control (M,N, n=4) and 25 pg hoxb4a RNA-injected (O,P, n=4) larvae were stained with anti-RFP (magenta) and anti-GFP (green) antibodies. (Q,R) Length of the gsx2:RFP+ hindbrain region (Q) and number of 28C;UAS:GFP+ neurons (R) in control and hoxb4a RNA-injected larvae. (S,T) Length of the gsx2:RFP+ hindbrain region in control (n=5), aldh1a2 morphant (n=8) and hoxb4a RNA-injected aldh1a2 morphant larvae (n=8) (S); in control (1% DMSO-treated, n=4), DEAB-treated (n=5), hoxb4a RNA-injected and DEAB-treated larvae (n=5) (T). (U,V) Number of 28C;UAS:GFP+ neurons in control (n=4), aldh1a2 morphant (n=4), and hoxb4a RNA-injected aldh1a2 morphant larvae (n=4) (U); in control (1% DMSO-treated, n=5), DEAB-treated (n=5), hoxb4a RNA-injected and DEAB-treated larvae (n=5) (V). *P<0.05; **P<0.01; ***P<0.001 (Student's t-test for K,L,Q,R, and one-way ANOVA with Tukey's multiple comparison test for S-V). Data are mean±s.e.m. with individual values indicated. Scale bars: 100 µm in A (applies to A-D), M (applies to M-P) and I (applies to I,J); 50 µm in E (applies to E,G); 20 μm in F (applies to F,H). PHENOTYPE:
|