- Title
-
BE4max and AncBE4max Are Efficient in Germline Conversion of C:G to T:A Base Pairs in Zebrafish
- Authors
- Carrington, B., Weinstein, R.N., Sood, R.
- Source
- Full text @ Cells
|
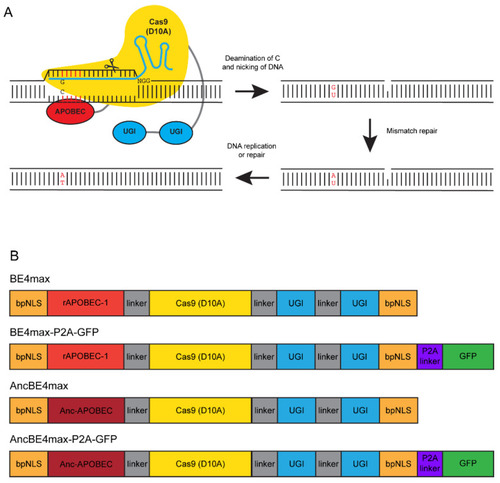
An overview of C → T conversion by BE4 and cytosine base editors used in this study. ( |
|
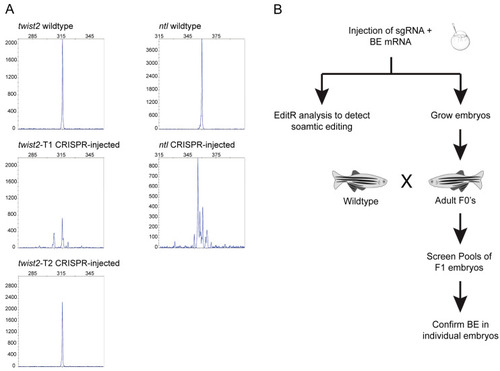
Selection of targets and strategy for assessment of base editing efficiency. ( |
|
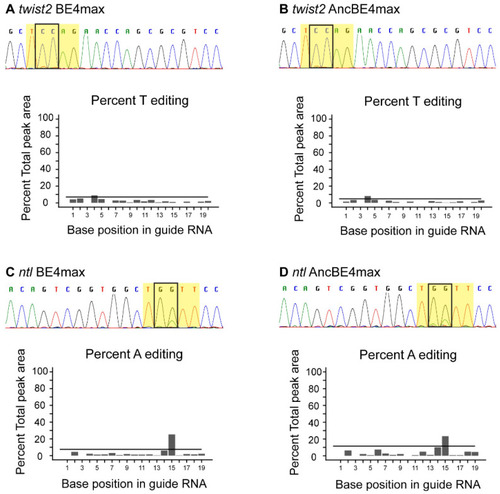
Somatic base editing with BE4max and AncBE4max. ( |
|
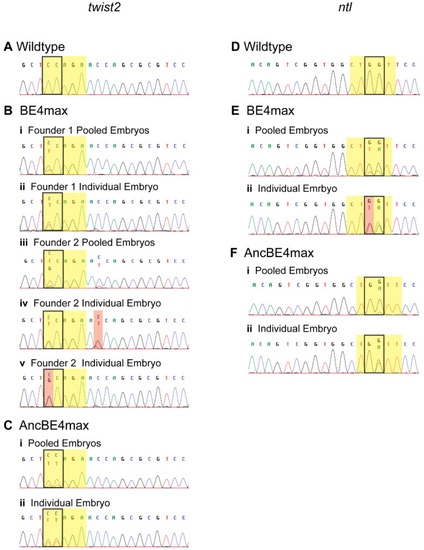
Germline base editing in pooled or individual embryos. Representative chromatograms of a wildtype embryo ( |