- Title
-
Klf9 is a key feedforward regulator of the transcriptomic response to glucocorticoid receptor activity
- Authors
- Gans, I., Hartig, E.I., Zhu, S., Tilden, A.R., Hutchins, L.N., Maki, N.J., Graber, J.H., Coffman, J.A.
- Source
- Full text @ Sci. Rep.
|
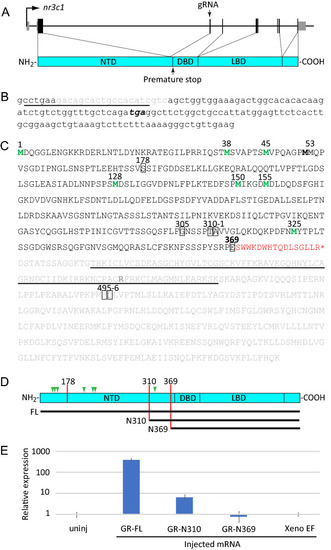
A new CRISPR-Cas9-induced mutation of the zebrafish GR |
|
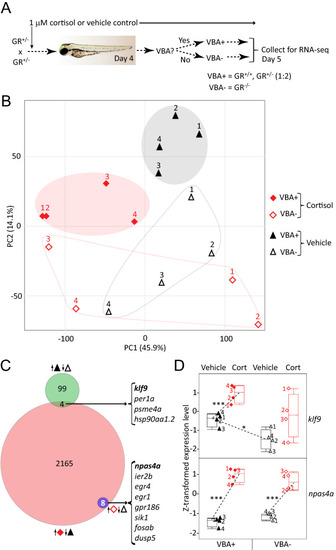
RNA-seq identifies GR-dependent and GR-independent genes and effects of chronic cortisol treatment. ( |
|
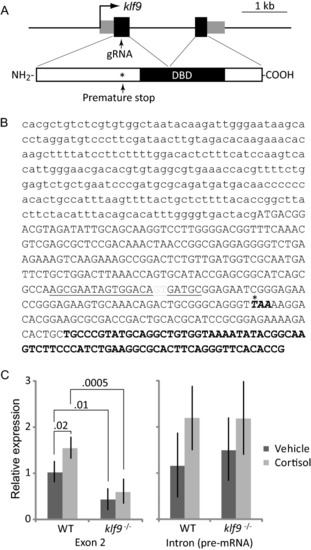
CRISPR-Cas9-mediated disruption of zebrafish |
|
RNA-seq identifies |
|
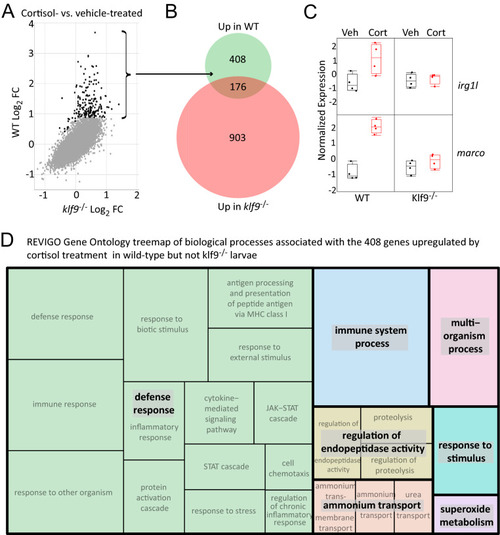
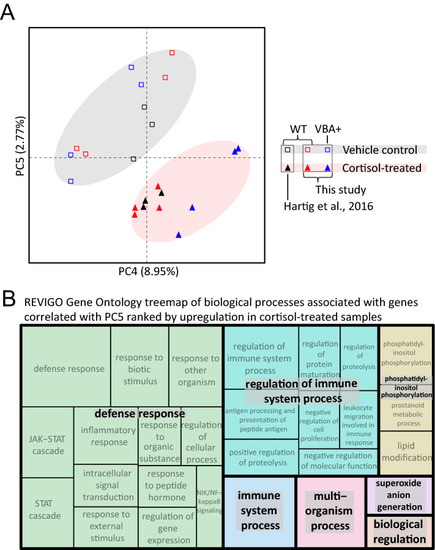
Combined analysis of three different RNA-seq datasets examining the transcriptomic effects of chronic cortisol exposure. ( |
|
Identification and analysis of a common set of genes upregulated by chronic cortisol treatment in multiple RNA-seq experiments. ( |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

Unillustrated author statements PHENOTYPE:
|