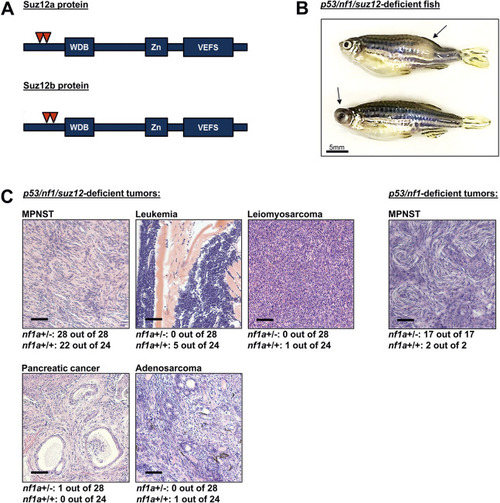
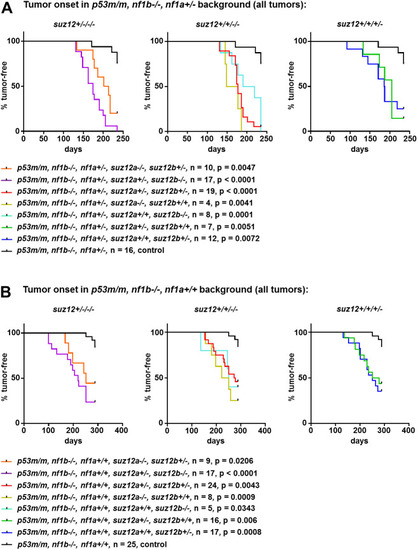
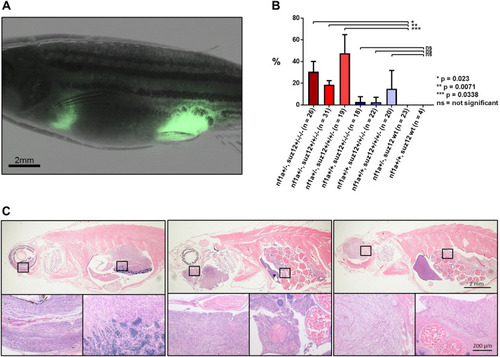
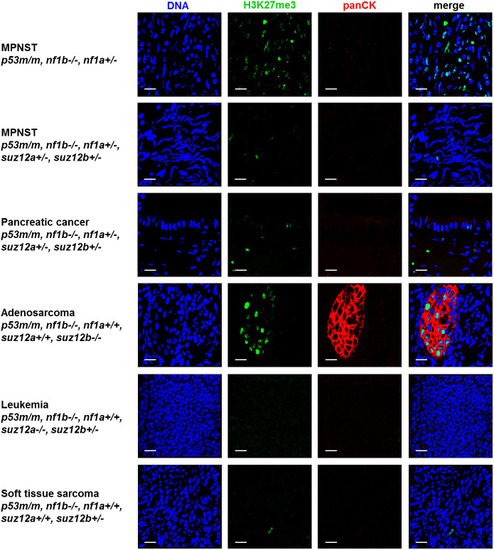
- Title
-
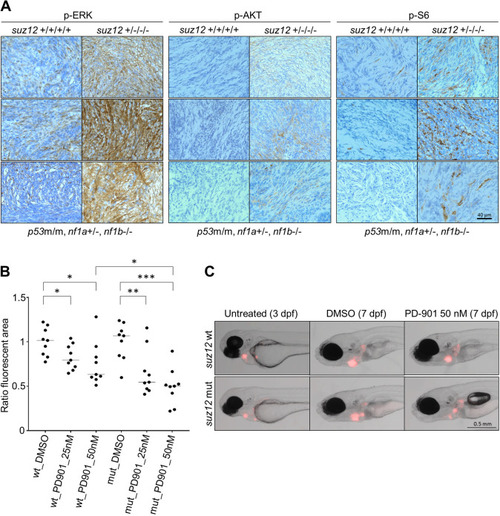
suz12 inactivation in p53 and nf1 deficient zebrafish accelerates the onset of MPNSTs and expands the spectrum of tumor types to include adenocarcinoma, leukemia, and soft tissue sarcoma
- Authors
- Oppel, F., Ki, D.H., Zimmermann, M.W., Ross, K.N., Tao, T., Shi, H., He, S., Aster, J.C., Look, A.T.
- Source
- Full text @ Dis. Model. Mech.
|
|
|
|
|
|
|
|
|
|