- Title
-
RNA exosome mutations in pontocerebellar hypoplasia alter ribosome biogenesis and p53 levels
- Authors
- Müller, J.S., Burns, D.T., Griffin, H., Wells, G.R., Zendah, R.A., Munro, B., Schneider, C., Horvath, R.
- Source
- Full text @ Life Sci Alliance

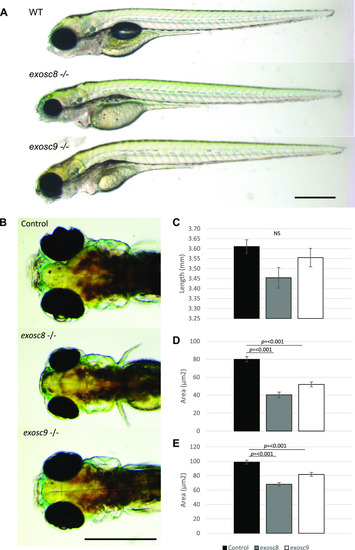
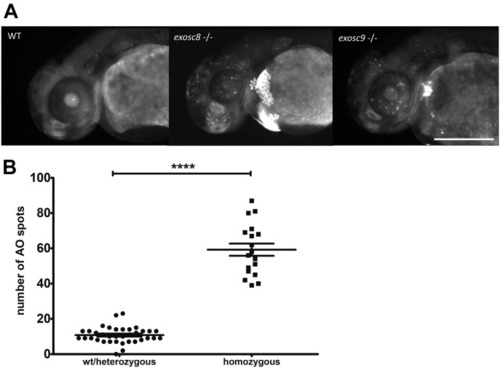
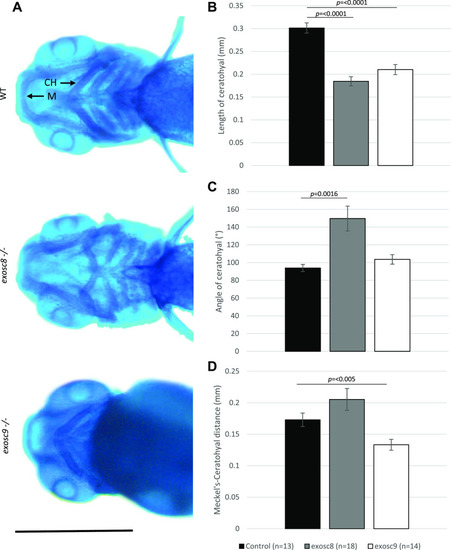
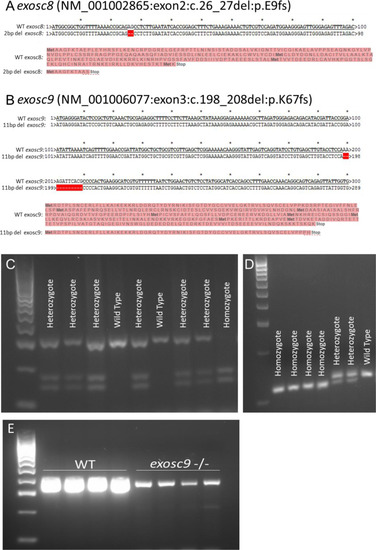
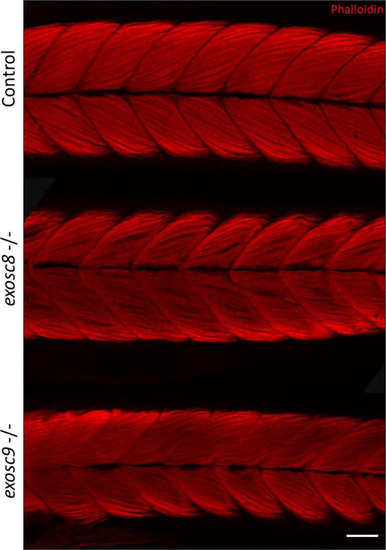
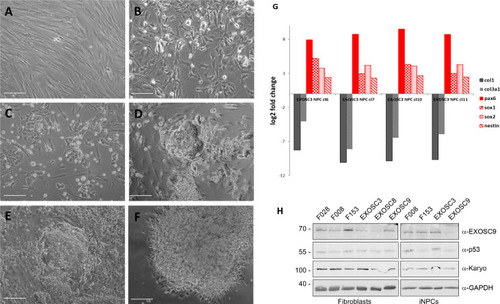
ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
|
|
PHENOTYPE:
|
|
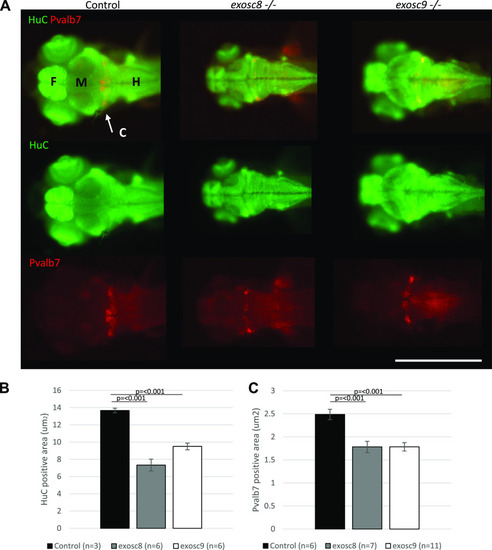
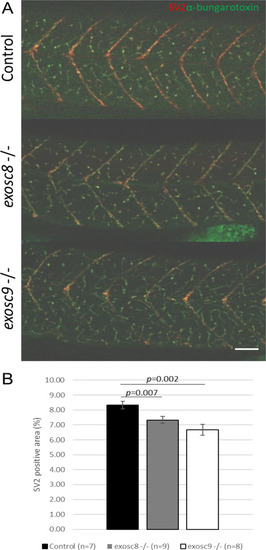
Immunofluorescence in 5-dpf wild-type, PHENOTYPE:
|
|
|