- Title
-
Chondroitin sulfate proteoglycan 4 regulates zebrafish body axis organization via Wnt/planar cell polarity pathway
- Authors
- Lee, Y.H., Kawakami, K., HuangFu, W.C., Liu, I.H.
- Source
- Full text @ PLoS One
|
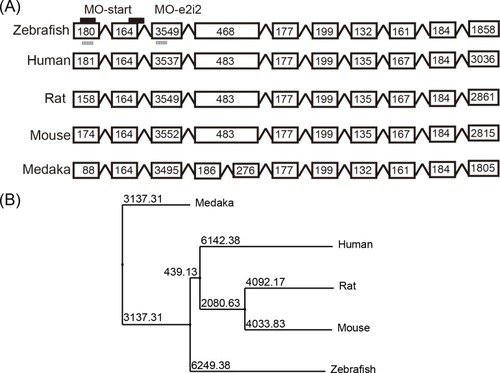
(A) The genome structure of |
|
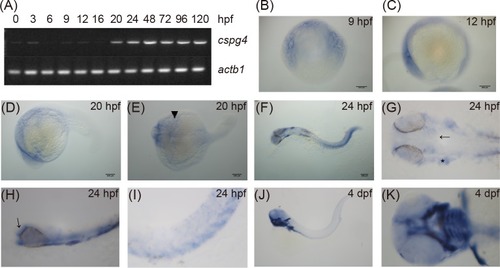
(A) The expression of |
|
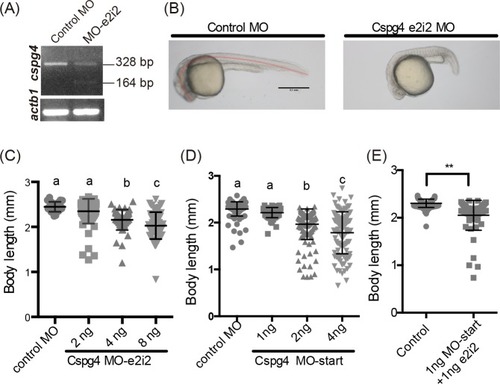
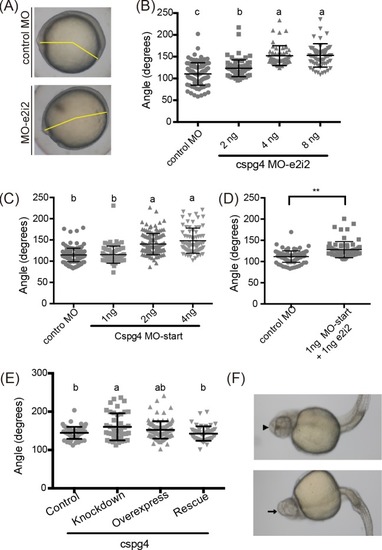
(A) The 164 bp band indicated the skipping of exon two. (B) PHENOTYPE:
|
|
The cartilage was stained by using Alcian blue staining. (A, B) The ventral view and lateral view of 4 ng control MO injected embryo at 3 dpf. (C, D) Four ng PHENOTYPE:
|
|
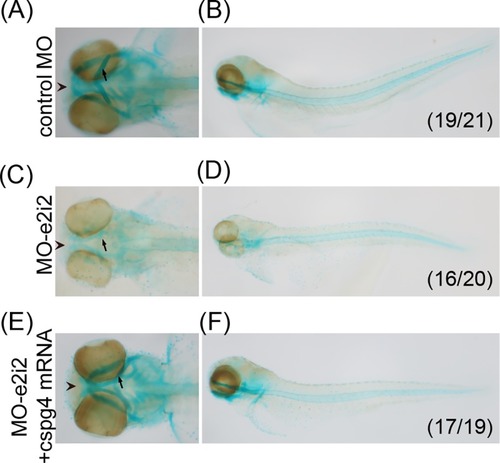
(A) Incomplete body axis organization was caused by PHENOTYPE:
|
|
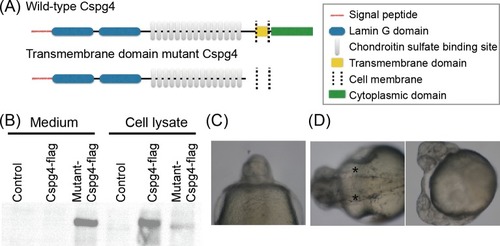
(A) Scheme illustrates wild-type and mutant Cspg4 protein structure. (B) The flag-tag signal of both wild-type and mutant Cspg4 were detected in the cell lysate, but in culture medium the signal of flag-tag was only detected in mutant Cspg4-flag transfected group. (C) Eight percent of PHENOTYPE:
|
|
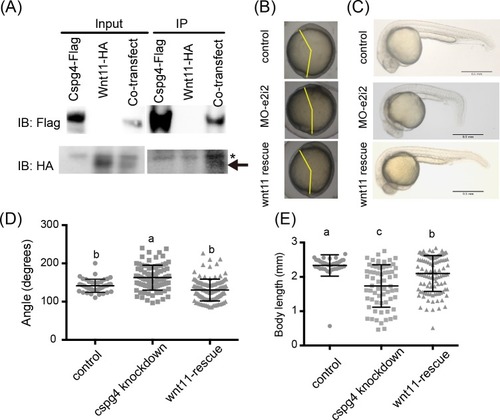
(A) Wnt11 had binding affinity with Cspg4. Wnt11-HA (indicated by arrow) was co-precipitated with Cspg4-flag. Asterisk mark (*) indicates non-specific band. (B) The abnormal body axis formation phenotype in |
|
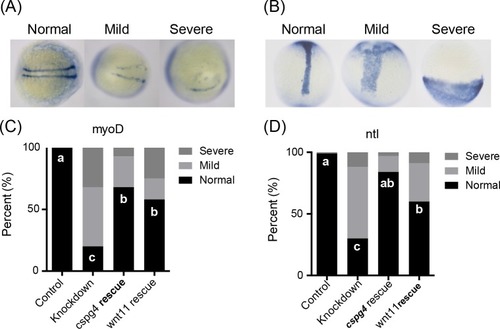
(A) EXPRESSION / LABELING:
PHENOTYPE:
|