- Title
-
Celsr1a is essential for tissue homeostasis and onset of aging phenotypes in the zebrafish
- Authors
- Li, C., Barton, C., Henke, K., Daane, J., Treaster, S., Caetano-Lopez, J., Tanguay, R.L., Harris, M.
- Source
- Full text @ Elife
|
( PHENOTYPE:
|
|
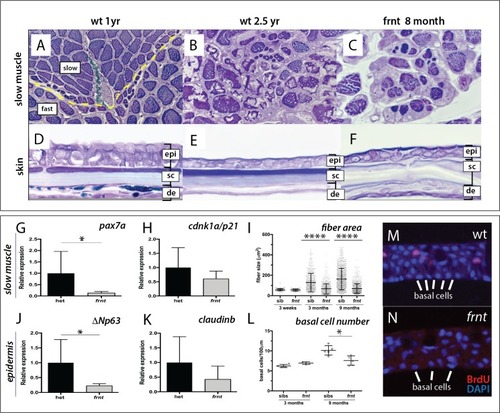
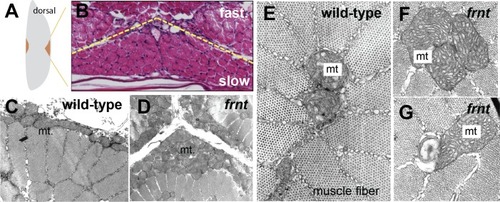
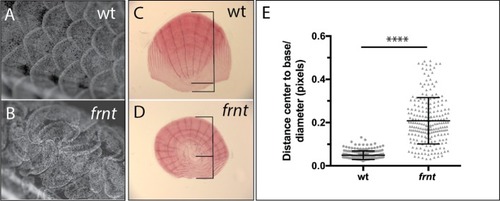
(A–F) Direct comparison of frnt with young adult, (A, D) and naturally aging wild-type zebrafish (2.5 years; (B, E). Similar degenerative pathologies are shared between aged zebrafish (B, E) and 8 month-old mutants (C, F), such as fibrosis and sarcopenia of slow muscle fibers (B, C), and thinning skin (E, F). (G–I) Characterization of slow muscle phenotype in frnt mutants. (G) Expression analysis of stem cell marker paired-box 7a (pax7a, n = 6) and (H) cdnk1a/p21 control in slow muscle from 7 month old homozygous (n = 6) and heterozygous mutant fish (n = 9). (I) Adult frnt has smaller fiber size in slow muscle compared to age-matched wild type and sibling fish (3 month (n = 5) and 9 month old (n = 9), but not as juvenile fish (3 week old, n = 3). (J–L) Changes in epidermal phenotype in frnt mutants. (J) Expression of the stem cell marker delta-Np63 and (K) control claudin-b in epidermal tissues from 7 month old homozygous (n = 12) and heterozygous (n = 5–6) frnt fish. (L) Count of DAPI positive basal cells in the integumentary epithelium in 3 (n = 3–4) and 9 month old (n = 5) frnt fish. (M–N). DAPI stained epidermal nuclei in wild-type (M) and frnt (N) mutant epidermis. Error represented as mean +/- standard deviation. ****p<0.0001, *p<0.05. EXPRESSION / LABELING:
PHENOTYPE:
|
|
( PHENOTYPE:
|
|
( PHENOTYPE:
|
|
( |
|
( |
|
( PHENOTYPE:
|
|
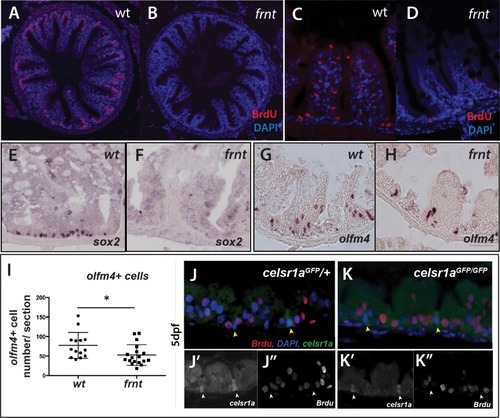
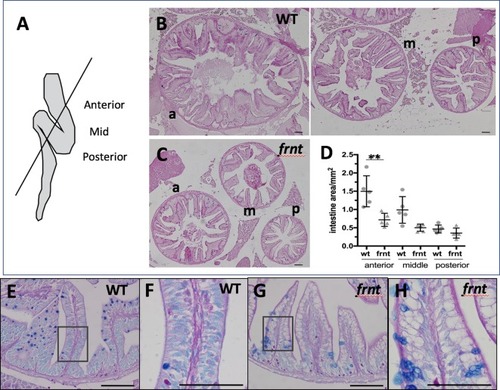
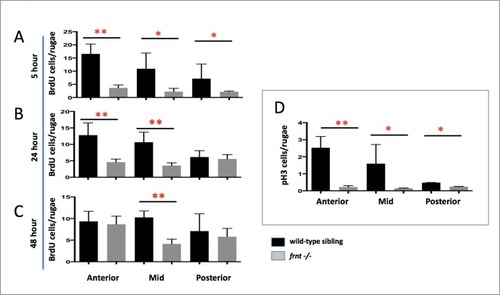
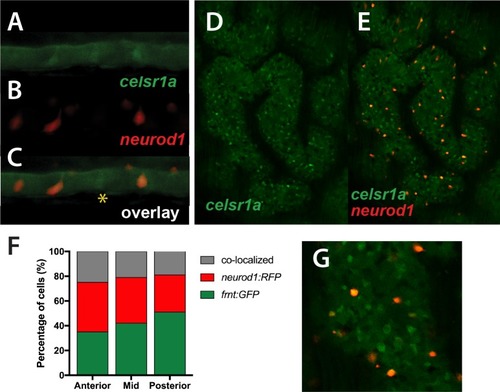
(A–D) Analysis of proliferative capacity of the adult intestine in celsr1a mutants and age matched wild-type fish (pulse injection and incorporation after 4 hr (red), nuclei counterstained by DAPI. (A, B) Low power view of comparable posterior regions of intestine of wild-type (A) and age and size matched frnt mutants (B). (C, D) Close up of intestinal rugae showing cells incorporating BrdU. (E–H), in situ hybridization of expression of sex-determining region Y-box 2 (sox2) (E, F) and olfactomedin 4 (olfm4). (G, H) genese in adult intestinal epithelia of wild-type (E, G) and celsr1a mutant (F, H) zebrafish. (I) Quantitation of changes in the number of olfm4+ cells observed in mutants; data presented as mean +/- standard deviation, *p<0.05, n = 5 (wt sibling), n = 7 (frnt) (J–K) Proliferative cells (24 hr after BrdU pulse, red) in comparison to celsr1a expression (green, yellow arrowhead) in larval developing intestine at 5dpf in (J) ceslr1aGFP heterozytotes and (K) ceslr1aGFP mutants; DAPI, blue. Insets J’ and J’’ and K’ and K’’are separate channels showing celsr1a expression and Brdu detection, respectively in the same tisse. |
|
( PHENOTYPE:
|
|
( |
|
( |
|
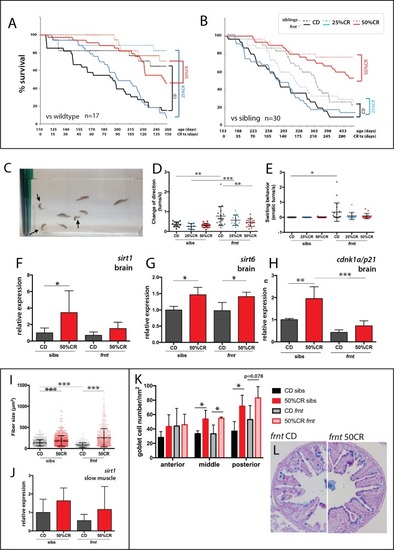
(A–C) Effects of specialized diets, having no restriction (control diet (CD), black), 25% (blue) or 50% (red) calorie restriction (CR), on viability of zebrafish; solid line, frnt mutant; dotted line, control fish. (A–B) The effect of the specific dietary reduction in calories on survival in frnt; 50%CR lead to significant increase in survival of mutant and siblings compared to control and 25%CR; differences between 50%CR diets for both mutant and siblings are all significant (p<0.001) compared to control and 25%CR by Mantel-Cox and Geha-Brelow-Wilcoxon tests. (C–E) Caloric restriction ameliorates aberrant swimming behavior in celsr1a mutants. (C) High frequency turning behavior of mutants in tank (arrows). Quantitation of change in direction (D) and erratic turns (E) in treatment groups with different levels of caloric reduction. (F–H) Expression of genes associated with senescence and lifespan in brain tissue of wild type and celsr1a mutants in different dietary treatments: (F) sirt1; (G) sirt6 and (H) cdnk1a/p21; data represented as mean ± standard deviation. (I–J) Effect of caloric restriction on slow muscle fiber area in wild-type and celsr1a mutant adult zebrafish. (I) Slow muscle fiber area is potentiated in response to 50%CR in both wild-type and mutants. (J) sirt1 expression in slow muscle from different diet treatment groups. (K–M), Effect of caloric restriction on intestinal differentiation phenotypes of wild-type celsr1a mutant adults. (K) goblet cell number quantitated from different regional areas of the gut in CD and 50%CR treatment groups. Data presented as mean + /- standard deviation, *p<0.05, paired t-test, n = 4 (frnt), n = 3 (sibs). (L) Goblet cell morphology in CD and 50%CR treated mutant posterior intestine. PHENOTYPE:
|
|
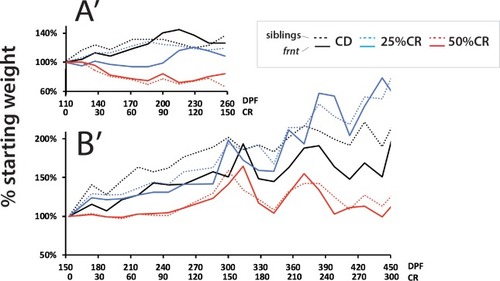
Analysis of average weight per treatment group for both experiments (A’ and B’, respective to experimental results in |
|
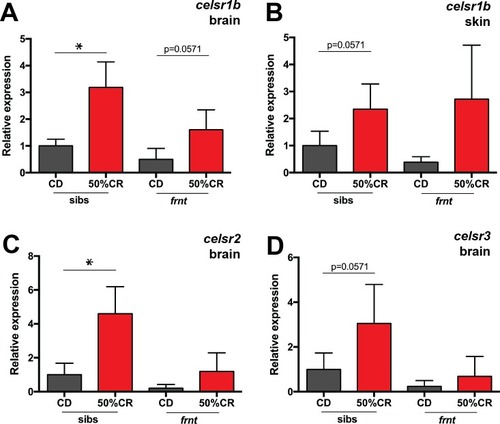
qRT-PCR analysis of |
|
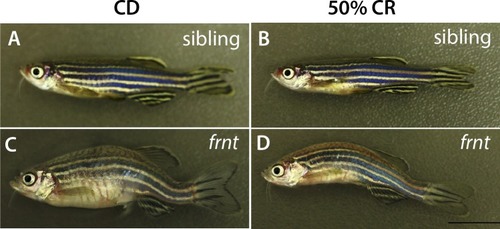
Representative adult fish after feeding regimen of control or calorie restricted (CR) diet. ( |

Unillustrated author statements PHENOTYPE:
|