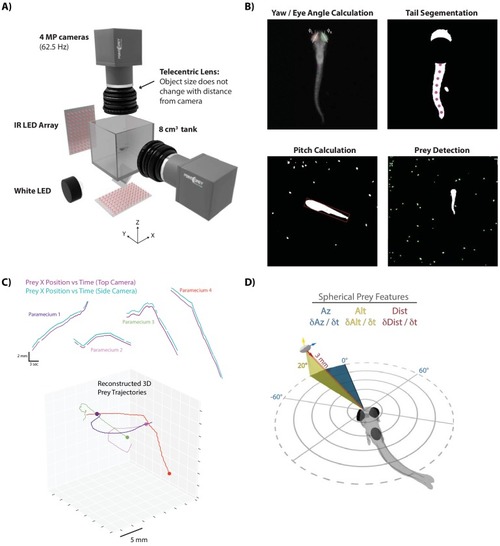
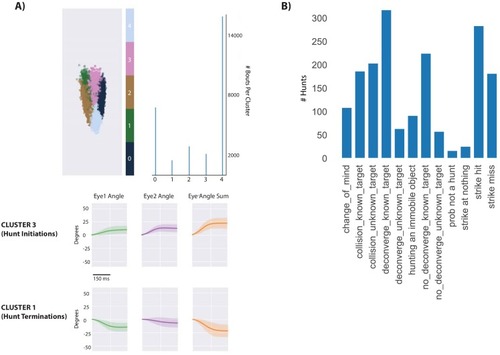
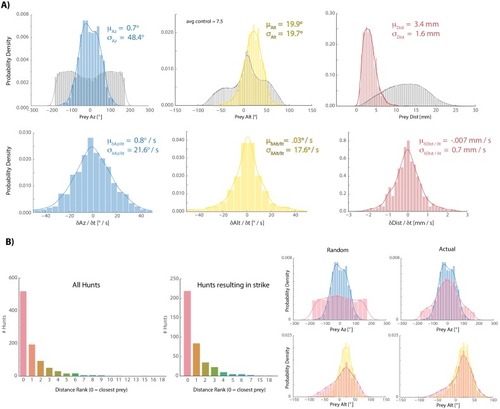
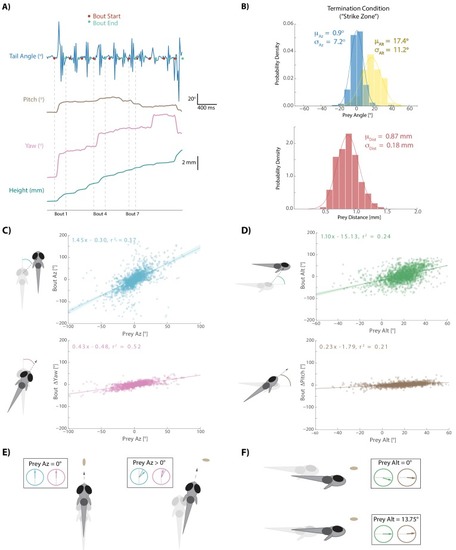
- Title
-
Elements of a stochastic 3D prediction engine in larval zebrafish prey capture
- Authors
- Bolton, A.D., Haesemeyer, M., Jordi, J., Schaechtle, U., Saad, F.A., Mansinghka, V.K., Tenenbaum, J.B., Engert, F.
- Source
- Full text @ Elife
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |
|
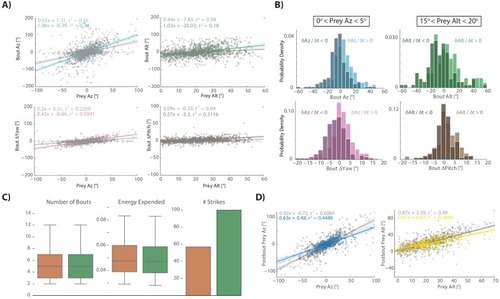
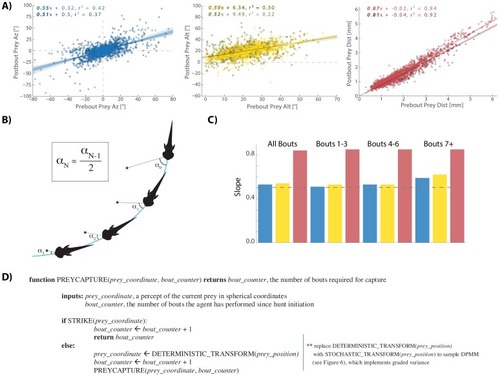
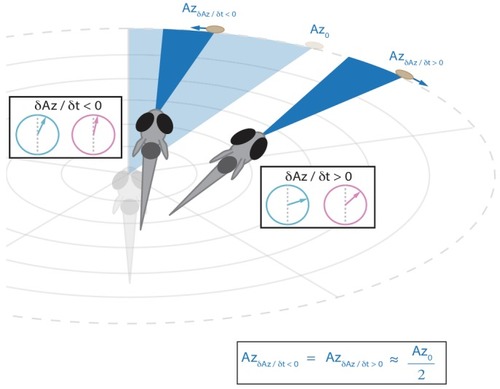
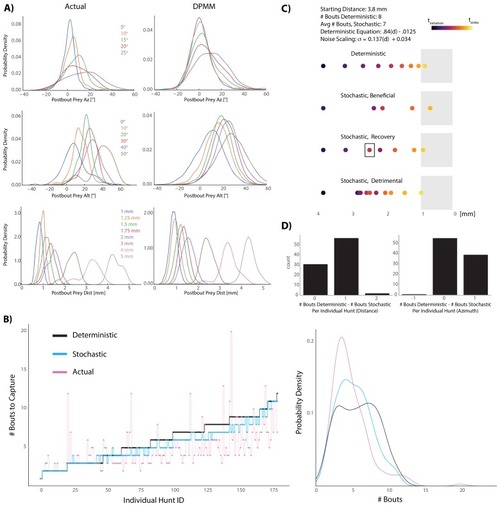
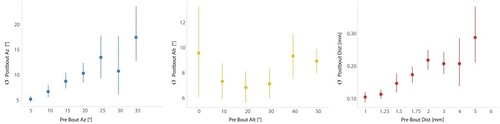
This strategy emerges from the position transforms in |
|
This strategy emerges from the position transforms in |
|
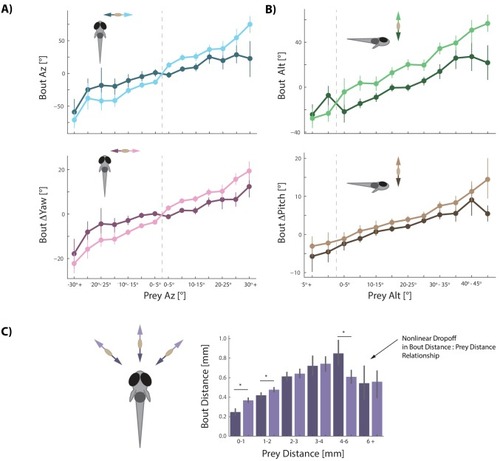
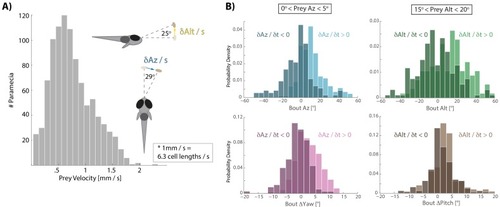
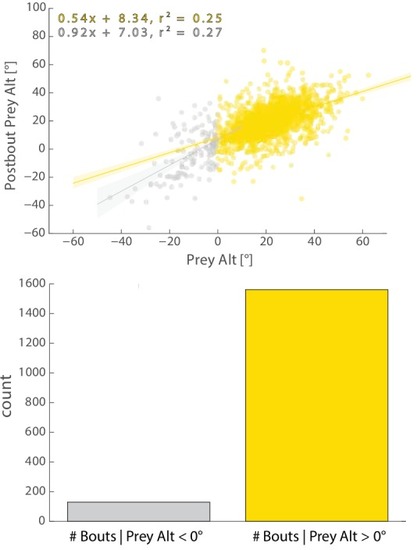
Most bouts in the dataset (>92%) occur when prey are above the fish. This change when prey cross 0° Alt is accounted for in all algorithms (see Appendix). |
|
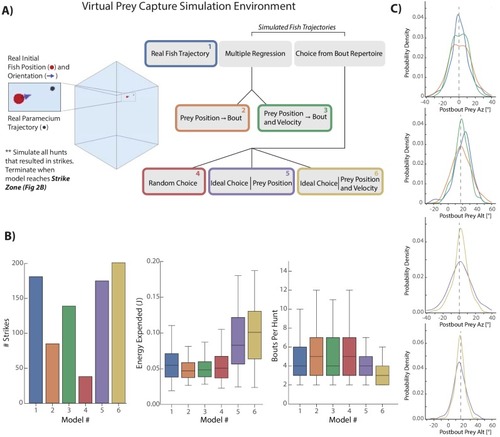
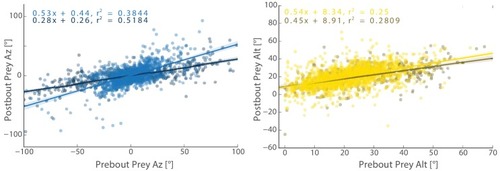
The initiation bout is a large angle turn that divides azimuth more than a pursuit bout; altitude is also significantly more reduced by the initiation bout. All regression models in simulations therefore use an independent regression fit to initiation bouts to start every simulated hunt. |
|
|
|
|