- Title
-
Lysosome-Rich Enterocytes Mediate Protein Absorption in the Vertebrate Gut
- Authors
- Park, J., Levic, D.S., Sumigray, K.D., Bagwell, J., Eroglu, O., Block, C.L., Eroglu, C., Barry, R., Lickwar, C.R., Rawls, J.F., Watts, S.A., Lechler, T., Bagnat, M.
- Source
- Full text @ Dev. Cell
|
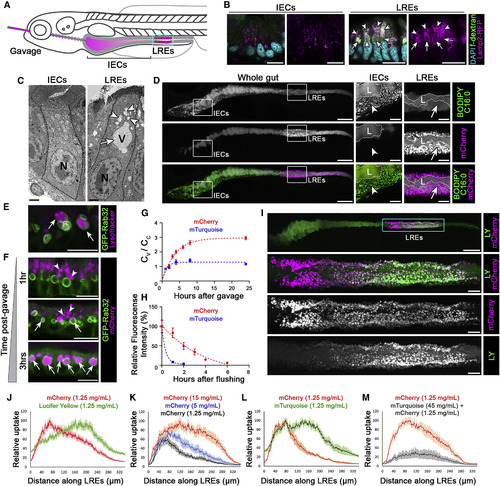
Luminal Proteins Are Internalized and Digested Intracellularly by LREs in the Zebrafish Intestine (A) Cartoon depicting gavage approach for studying luminal cargo uptake into LREs (cyan box) in the larval zebrafish intestine. (B) Confocal images of cross sections of intestinal segments showing IECs and LREs from a 6 dpf larva following gavage with fluorescent dextran (fDex). Lamp2 is highly expressed in LREs and localizes to endosomes (arrowheads) and lysosomal vacuoles (arrows) that accumulate internalized fDex. n ≥ 10 animals. Scale bars, 10 μm. (C) EM images of an IEC and an LRE. Arrowheads mark endosomes and an arrow the lysosomal vacuole. N = nucleus; V = vacuole. n ≥ 5 animals. Scale bars, 2 μm. (D) Live confocal images of a 6 dpf larval zebrafish intestine following gavage and internalization of BODIPY C16:0 and mCherry. IECs take lipids and LREs protein. L = lumen. Insets show magnified images marking lipid droplets (arrowheads) in IECs and lysosomal vacuoles in LREs (arrows). n = 5. Scale bars, 100 μm (whole gut images), 25 μm (magnified images). (E) Live confocal images of LREs of 6 dpf larva expressing GFP-Rab32a 1 h after gavage with LysoTracker, which labels vacuoles (arrows). n ≥ 10 animals. Scale bar, 10 μm. (F) Live confocal images of LREs of 6 dpf larvae expressing GFP-Rab32a that were gavaged with mCherry. Luminal protein (mCherry) was internalized, migrated through apical endosomes (arrowheads), and progressively accumulated in LRE vacuoles (arrows). Scale bars, 10 μm. (G) Quantitation of mCherry and mTurquoise accumulation in LRE vacuoles (Cv) respect to total cellular mean pixel intensity (Cc) over time after gavaging with 1.25 mg/mL of mCherry or mTurquoise. Data are fitted with the model Cv/Cc = k_in/k_ex ∗(1−exp(−k_ex ∗ X)) where k_in is the vacuole internalization rate and k_ex is the degradation rate. n ≥ 28; LREs from at least 6 animals per timepoint. (H) Quantitation of mCherry and mTurquoise degradation in LREs over time following gavage with 1.25 mg/mL mCherry or 5 mg/mL mTurquoise for 1 h. Data are fitted with one phase decay model (dotted lines). n ≥ 9 animals per timepoint. (I) Live confocal image of a 6 dpf larva gavaged with lucifer yellow (LY) and mCherry. Cyan box indicates magnified inset of LREs in lower panels. Scale bar, 100 μm (whole gut image), 20 μm (magnified images). (J) Internalization profiles of LY and mCherry along LREs (n = 12). (K) Internalization profiles along LRE region following gavage with increasing concentrations of mCherry: 1.25 mg/mL (n = 7), 5 mg/mL (n = 8), 15 mg/mL (n = 9). (L) Internalization profiles along LRE region of mCherry and mTurquoise (n = 8). (M) Internalization profiles along LRE region of mCherry following gavage with either mCherry alone (n = 7) or both mCherry + excess mTurquoise to outcompete mCherry uptake (n = 7). Data are means ± S.E.M. in (G), (H), and (J–M). Whole gut images in (D) and (I) are digitally stitched. See also Figures S1–S3. |
|
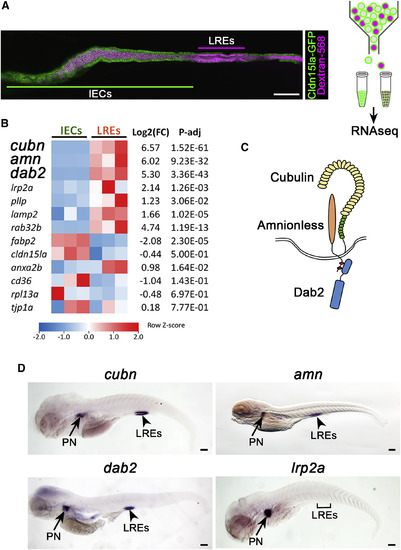
LREs Specifically Express a Multi-ligand Endocytic Machinery Composed of Cubn, Amn, and Dab2 (A) Isolation and analysis of gene expression profiles in LREs versus IEC. 6 dpf larvae expressing Cldn15la-GFP throughout the gut were gavaged with alexa-568 dextran (left panel) and subjected to FACS to isolate IECs and LREs for RNA-seq (right panel). Scale bar, 100 μm. (B) Heatmap of RNA-seq data depicting expression levels of selected genes in LREs and IECs. Three independent samples for each cell population were used for RNA-seq experiment. (C) Cartoon illustrating the multi-ligand endocytic machinery composed of Cubn, Amn, and Dab2. (D) In situ hybridization of cubn (cubilin), amn (amnionless), dab2, and lrp2a (megalin). cubn, amn, and dab2 transcripts are enriched in LREs and pronephros. Bracket marks LRE region devoid of lrp2a transcript expression. PN = pronephros. n ≥ 20 animals, Scale bar, 100 μm. See also Figures S4 and S5. |
|
The Cubn/Amn Endocytic Receptor Complex Mediates Protein Uptake in LREs (A) Live confocal images showing LY and mCherry uptake in LREs of cubnpd1169 +/+, +/−, −⁄− 6 dpf larvae. Larvae were imaged 5 h after gavage. cubn pd1169 mutants show selective loss of protein (mCherry) internalization. Scale bar, 50 μm. (B and C) Internalization profiles of LY (left) and mCherry (right) of 6dpf cubn pd1169 +/+ (n = 5), +/− (n = 10), −⁄− (n = 9) (B) and of amn pd1189+/+ (n = 4), +/− (n = 15), −⁄− (n = 4) (C). Endocytic receptor mutants cubnand amn exhibited specific defects in protein uptake (arrows). Data are means ± S.E.M. See also Figures S5 and S6. PHENOTYPE:
|
|
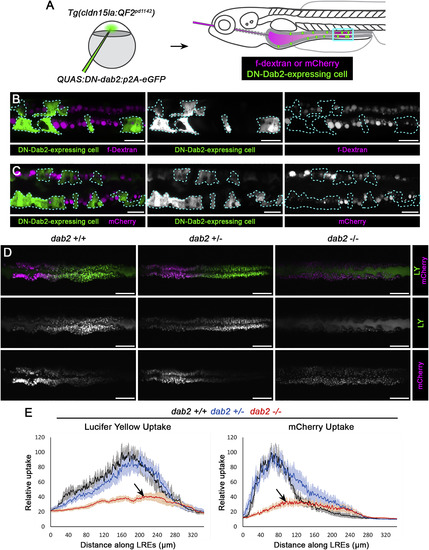
Dab2 Mediates Fluid-Phase and Receptor-Dependent Endocytosis in LREs (A) Experimental scheme to mosaically inhibit Dab2 function. A dominant-negative (DN), QUAS:DN-Dab2:p2A-EGFP, construct was injected into Tg(cldn15la:QF2pd1142) embryos to co-express DN-Dab2 and soluble eGFP to mark expressing cells in IECs and LREs. Larvae mosaically expressing DN-Dab2 were then gavaged with fDex or mCherry and imaged 5 h post-gavage. (B and C) Live confocal images of fDex or mCherry uptake in 6 dpf Tg(cldn15la:QF2 pd1142) LREs mosaically expressing DN-Dab2. LREs expressing DN-Dab2 (marked with dotted line) showed reduced fDex (n = 6) (B) and mCherry (n = 3) (C) uptake relative to WT neighboring LREs. Scale bar, 20 μm. (D) Live confocal images of 6 dpf dab2pd1162 +/+, +/−, −⁄− LREs imaged 5 h after gavage with LY and mCherry. Scale bar, 50 μm. (E) Internalization profiles of LY (left) and mCherry (right) from 6 dpf dab2 pd1162 +/+ (n = 5), +/− (n = 8), −⁄− (n = 6) larvae. dab2 mutants showed impaired internalization of both protein (mCherry) and fluid-phase (LY) cargos (arrows). Data are means ± S.E.M. Images in (C) are digitally stitched. See also Figures S5 and S6. PHENOTYPE:
|
|
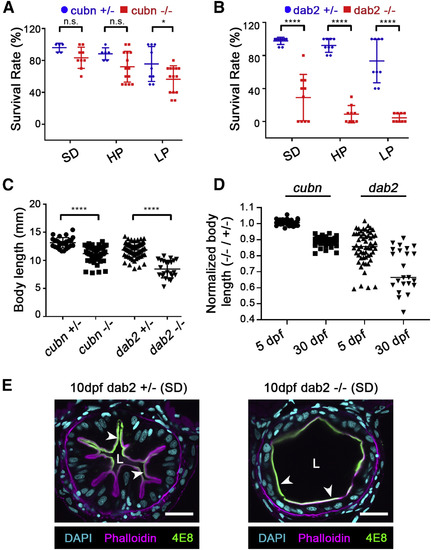
Loss of LRE Protein Uptake Impairs Growth and Survival in Larval Zebrafish (A and B) Survival rates of cubn pd1169 +/−, −⁄− (A), and dab2 pd1162 +/−, −⁄−(B) 30 dpf larvae raised under different feeding conditions: non-calorie restricted standard diet (SD), calorie-restricted high-protein diet (HP) and calorie-restricted low-protein diet (LP). n = 5, 9, 6, 14, 9, and 14 (A; left to right), n = 9, 10, 9, 9, 9, and 9 (B; left to right). Data points are the survival rates of each tank subjected to the experiment. (C) Comparison of body lengths of cubn pd1169 +/− (n = 29), −⁄− (n = 57), and dab2 pd1162 +/− (n = 69), −⁄− (n = 23) 30 dpf larvae raised under a non-calorie restricted SD. Data points are the body lengths of individual 30 dpf larvae that survived until the end of the feeding experiments. (D) Quantitation of mutant/heterozygous control body length ratios for cubn pd1169 and dab2 pd1162 larvae before onset of feeding (5 dpf) and after being fed with a non-calorie restricted SD (30 dpf). Growth deficits largely occur post-embryonically. (E) Intestine cross section images of 10 dpf dab2 pd1162 +/− or −⁄− larvae raised under non-calorie restricted SD. Arrowheads point to 4E8 colocalization with phallodin, indicating no polarization defect in some of the dab2 −⁄−. L = lumen. n ≥ 15 animals/genotype. Scale bar, 20 μm. ∗p < 0.05, ∗∗∗∗p < 0.0001; two-tailed unpaired t tests (A–C). p values, t values and degree of freedom for the statistical tests are provided in the Table S3. PHENOTYPE:
|
|
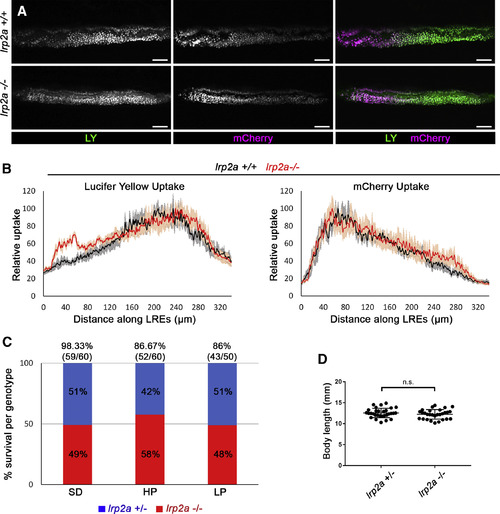
lrp2a (megalin) Mutants Do Not Show Defects in LRE Uptake or Survival under Different Feeding Conditions (A) Live confocal images showing LY or mCherry uptake in LREs of lrp2amw1 +/+, −⁄− 6 dpf larvae. The larvae were imaged 5 h after gavage. Scale bar, 20 μm. (B) Internalization profiles of LY (left) and mCherry (right) along LREs of 6 dpf lrp2amw1 +/+ (n = 5), −⁄− (n = 5). Data are means ± S.E.M. (C) Survival rates of lrp2amw1 +/− and −⁄− 30 dpf larvae raised under different feeding conditions: non-calorie restricted standard diet (SD), calorie-restricted high-protein diet (HP), and calorie-restricted low-protein diet (LP). Larvae from the cross lrp2amw1 +/− X lrp2amw1 −⁄−were used for the experiment. At 30 dpf, the survived larvae were counted and genotyped to determine what percentage of survived larvae are lrp2a +/− or −⁄−. The numbers above the graph bars indicate the percentage of overall tank survival for each feeding condition. (D) Comparison of body lengths of lrp2amw1 +/− (n = 30) and −⁄− (n = 29) 30 dpf larvae raised under non-calorie restricted SD. Two-tailed unpaired t test was used for statistical analysis. p values, t values and degree of freedom for the statistical tests are provided in the Table S3. PHENOTYPE:
|
|
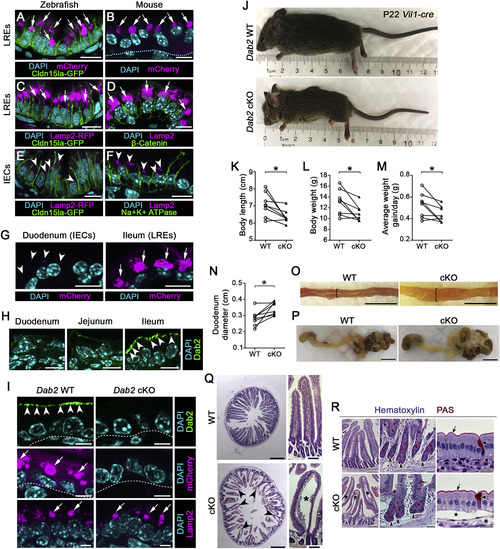
Loss of LRE Function Results in Growth Stunting and Intestinal Swelling in Suckling Mice (A and B) Confocal images of LREs of TgBAC(cldn15la-GFP pd1034)zebrafish larva (A) and of P7 neonatal mouse (B) after mCherry uptake. Arrows point to lysosomal vacuoles filled with mCherry. Dashed line in (B) indicates basement membrane. Scale bar, 10 μm. (C–F) Confocal images of LREs (C and D) and IECs (E and F) of TgBAC(cldn15la-GFP pd1034); TgBAC(lamp2-RFP pd1044) zebrafish larva and P7 neonatal mouse intestine stained with LAMP2 antibody, showing the presence of lysosomal vacuoles (arrows) in LREs (C and D) and lysosomes (arrowheads) in IECs (E and F). Scale bar, 10 μm. (G) Confocal images of duodenum and ileum of P6 neonatal mouse after mCherry uptake. Arrowheads point to small vesicular structures with mCherry in IECs and arrows point to lysosomal vacuoles filled with mCherry in LREs. Scale bar, 10 μm. (H) DAB2 expression in the duodenum, jejunum, and ileum of P3 WT neonatal mouse. DAB2 expression is enriched in the ileum (arrowheads), the segment harboring LREs. Scale bar, 10 μm. (I) mCherry uptake in the ileum of P6 Dab2 WT and Dab2 cKO (Vil1-Cre;Dab2tm1Cpr fl/fl) mice. While DAB2 expression (top images; arrowheads) and mCherry protein uptake (middle images; arrows) is abolished in Dab2 cKO mouse ileal LREs, LAMP2 expression is not (bottom images; arrows). Scale bar, 5 μm. (J) Images of P22 Dab2 WT and Dab2 cKO mice. (K–N) Quantitation of body length (K), weight (L), average weight gain/day (M), and duodenum diameter (N) of P22 Dab2 WT and cKO mice. (O) Images of dissected duodenum of P22 Dab2 WT and cKO mice. Brackets show prominent swelling of the GI tract in Dab2 cKO mice. Scale bar, 1 cm. (P) Images of the dissected stomach and intestinal tract of WT and Dab2 cKO mice harvested at P22. Scale bar, 1 cm. (Q) Images of H&E stained sections of the duodenum of P22 Dab2 WT and cKO mice, which exhibit severe distended villi (arrowheads and asterisk) indicative of submucosal edema. Scale bar, 500 μm (transverse cross sections), 100 μm (longitudinal cross sections). (R) Hematoxylin and periodic acid–Schiff (PAS) staining of duodenal sections of WT and Dab2 cKO P22 mice. Concave arrowheads point to Paneth cells within crypts; flat arrowheads show goblet cells; arrows mark apical membrane; asterisks indicate edema. Scale bar, 200 μm (left), 40 μm (middle), and 20 μm (right). In (K–N), data points are littermate averages for each genotype and littermate averages of WT and cKO from the same litter are paired with lines. Measurements of individual mice from 8 different litters graphed in (K–N) are provided in the Table S2. ∗p < 0.05; Two-tailed paired t tests (K–N). p values, t values, and degree of freedom for the statistical tests are provided in Table S3. |
Reprinted from Developmental Cell, 51(1), Park, J., Levic, D.S., Sumigray, K.D., Bagwell, J., Eroglu, O., Block, C.L., Eroglu, C., Barry, R., Lickwar, C.R., Rawls, J.F., Watts, S.A., Lechler, T., Bagnat, M., Lysosome-Rich Enterocytes Mediate Protein Absorption in the Vertebrate Gut, 7-20.e6, Copyright (2019) with permission from Elsevier. Full text @ Dev. Cell