- Title
-
Yap1b, a divergent Yap/Taz family member, cooperates with yap1 in survival and morphogenesis via common transcriptional targets
- Authors
- Vázquez-Marín, J., Gutiérrez-Triana, J.A., Almuedo-Castillo, M., Buono, L., Gómez-Skarmeta, J.L., Mateo, J.L., Wittbrodt, J., Martínez-Morales, J.R.
- Source
- Full text @ Development
|
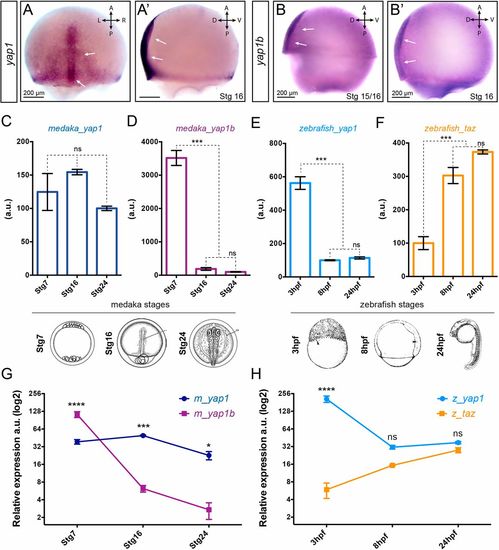
Expression analysis of Yap family genes. In situ hybridization analysis of yap1 (A,A′) and yap1b(B,B′) distribution in stage 15-16 medaka embryos. Scale bars: 200 µm. Quantification of the expression levels of Yap family genes at three equivalent developmental stages (i.e. early morula, late gastrula and optic cup stages) in medaka (C,D) and zebrafish (E,F) embryos by qPCR-RT (n=3). Relative expression level between paralogous genes was determined in medaka (G) and zebrafish (H) samples by droplet digital PCR (ddPCR) (n=2). One-way ANOVA analysis followed by Fisher's LSD test was used to evaluate statistical significance. ****P<0.0001; ***P<0.001; *P<0.05; ns, not significant. |
|
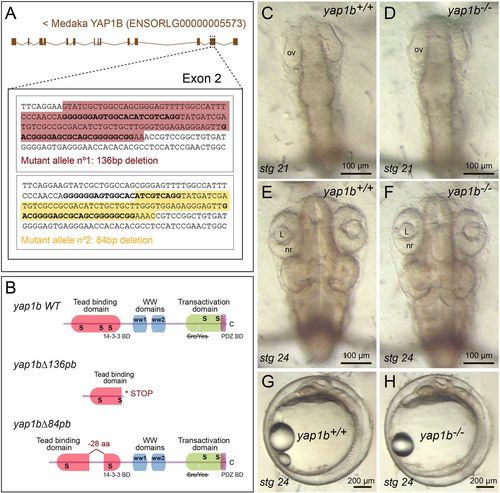
Generation of yap1b mutant alleles by CRISPR/Cas9 genome editing. (A) Schematic representation of the 136 bp and 84 bp deletions (shadowed sequence) generated in exon 2 of yap1b in medaka (see Materials and Methods). (B) The predicted Yap1b protein structure is depicted for the wild-type and mutant proteins. The yap1bΔ136pb deletion results in a frameshift mutation and premature stop codon, whereas yap1bΔ84pb results in a 28 amino acid deletion of the TEAD-binding domain. No phenotypic differences were observed when comparing wild-type (C,E,G) and yap1b mutant embryos (D,F,H) at stages 21 (C,D) and 24 (E-H). ov, optic vesicle; L, lens; nr, neural retina. |
|
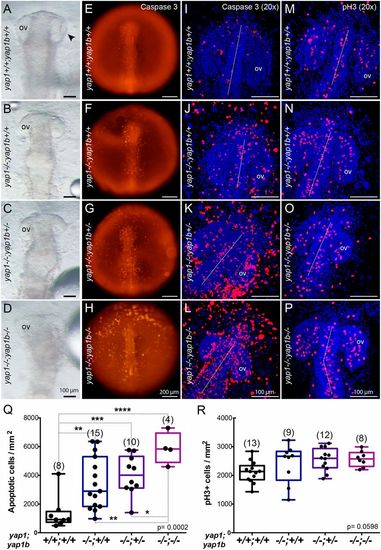
yap1 and yap1b cooperate in regulating cell survival and tissue morphogenesis. Bright-field images of the anterior half of yap1+/+;yap1b+/+ (A), yap1−/−;yap1b+/+ (B), yap1−/−;yap1b+/− (C) and yap1−/−;yap1b−/− (D) stage 21 embryos from a double heterozygous cross showing negative epistatic interaction between yap1 and yap1b. yap1−/− enhanced malformations in yap1−/−;yap1b−/− double mutants (D). Caspase 3 staining reveals apoptotic cells in stage 20 yap1+/+;yap1b+/+ (E,I), yap1−/−;yap1b+/+ (F,J), yap1−/−;yap1b+/− (G,K) and yap1−/−;yap1b−/−embryos (H,L). Whole embryos are shown under the fluorescent stereo microscope (E-H). DAPI counterstained confocal images show individual apoptotic cells (I-L). DAPI counterstained confocal images of pH3-positive mitotic cells in stage 20 yap1+/+;yap1b+/+ (M), yap1−/−;yap1b+/+(N), yap1−/−;yap1b+/− (O) and yap1−/−;yap1b−/− embryos (P). Quantification of caspase 3-positive cells per area shows a significantly increased apoptosis with fewer functional copies available of the Yap paralogs (Q). Quantification of pH3-positive cells per area shows not significant differences in mutants for the Yap paralogs (R). Boxes represent the quartiles; the whiskers indicate the the maximum and minimum values. One-way ANOVA analysis followed by Fisher's LSD test was used to evaluate statistical significance. ov, optic vesicle. ****P<0.0001; ***P<0.001; **P<0.01; *P<0.05. |
|
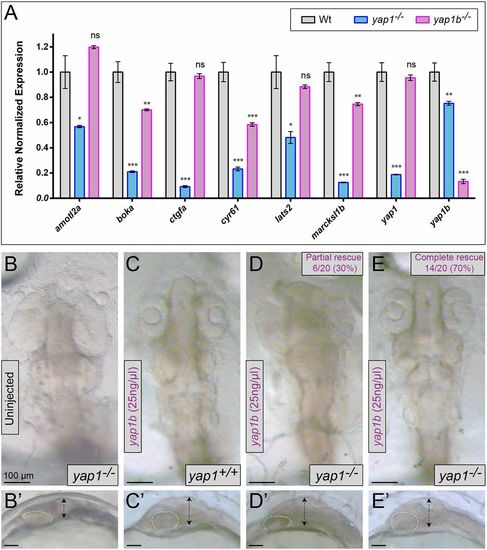
yap1/yap1b relative regulatory weight and yap1b rescuing capacity. (A) Quantification of the expression levels of genes targeted by Yap family proteins in wild-type, yap1−/− and yap1b−/−medaka embryos (stage 16) by qPCR (n=3). Comparative dorsal (B-E) and lateral (B′-E′) views of stage 24 medaka embryos in yap1b rescuing experiments. Embryo genotype (either yap1+/+ or yap1−/−) and yap1b injections are indicated. Embryo flattening (double-headed arrows) and optic cup shape (dotted lines) are also highlighted. The complete yap1−/− phenotypic rescue upon yap1b mRNA injection (E). One-way ANOVA analysis followed by Fisher's LSD test was used to evaluate statistical significance. ***P<0.001; **P<0.01; *P<0.05; ns, not significant. |
|
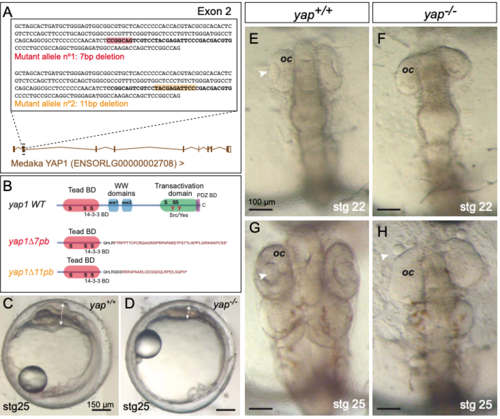
Generation of yap1 mutant alleles by CRISPR/Cas9 genome editing. (A) Schematic representation of the 7 bp and 11 bp deletions generated in the exon 2 of yap1 in medaka. The sequence in bold corresponds to the region targeted by two partially overlapping sgRNAs (see Methods). The boxes in red and orange correspond to the 7 and 11 bp deletions generated. (B) The predicted yap1 protein structure is depicted for the wild type and mutant proteins. In both cases, the deletions resulted in frame shift mutations and premature stop codons. (C-H) Phenotypic analysis of wild type (C, E, G) and yap1 mutant embryos (D, F, H) at stages 22 (E-F) and 25 (C-D, G-H) shows characteristic malformations, such as optic cup flattening and lens misalignment (arrows in H). oc = optic cup. Magnification bars are included in the figure. |
|
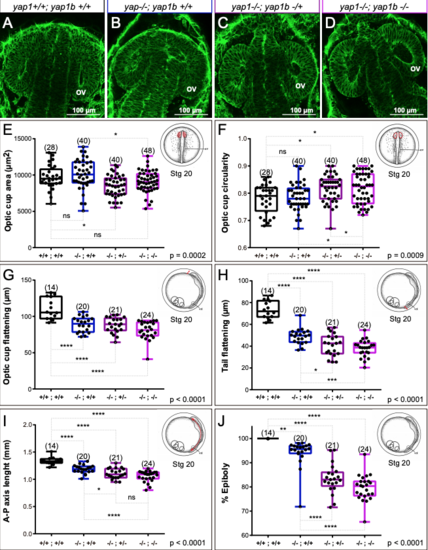
Quantitative analysis of morphogenetic defects in yap1/yap1b single and double mutants. (A-D) Confocal sections of phalloidin stained embryos showing optic cup morphology in wild type, yap1 mutants, and yap1/yap1b double mutants. (E-J) Quantitative analysis of optic cup area (E), circularity (F), and flattening (G); as well as tail flattening (H), embryo axis lenght (I), and % of epiboly (J) in wild type and mutant embryos reveal enhanced tissue malformations in yap1/yap1b double mutants. One Way ANOVA analysis followed by Fisher's LSD test was used to evaluate statistical significance. ov = optic vesicle. Magnification bars are included in the figure. |