- Title
-
Precise A•T to G•C base editing in the zebrafish genome
- Authors
- Qin, W., Lu, X., Liu, Y., Bai, H., Li, S., Lin, S.
- Source
- Full text @ BMC Biol.
|
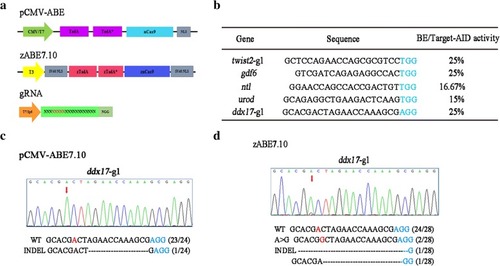
Comparison of the original ABE system and optimized zABE7.10 system in zebrafish. |
|
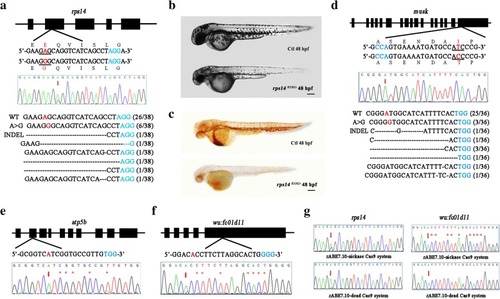
Codon-optimized zABE7.10 system in zebrafish induces A to G base conversion in zebrafish. |
|
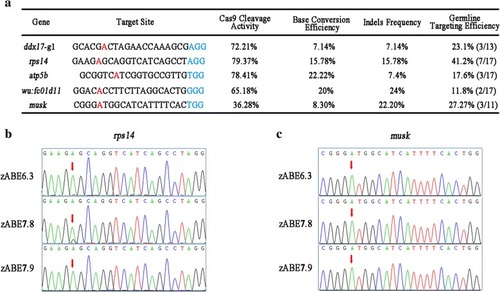
Summary of adenine base-editing results in zebrafish. |
|
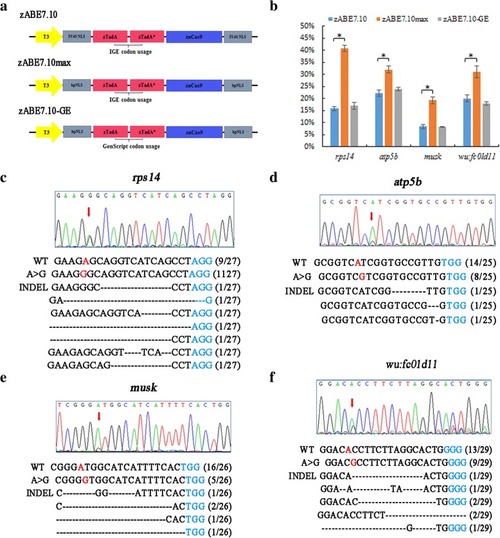
Adenine base editors modified by bpNLS enhance the base editing efficiency in zebrafish. |