- Title
-
Eaf1 and Eaf2 mediate zebrafish dorsal-ventral axis patterning via suppressing Wnt/β-Catenin activity.
- Authors
- Liu, J.X., Xu, Q.H., Yu, X., Zhang, T., Xie, X., Ouyang, G.
- Source
- Full text @ Int. J. Biol. Sci.
|
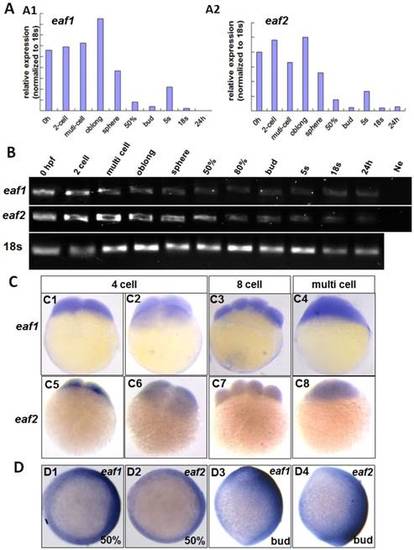
Maternal expression of eaf1 and eaf2 was detected by qRT-PCR (A), semi quantitative PCR (B), and whole mount in situ hybridization (C). (A, B) Embryos from 0 hpf, 2-cell stage (2-cell), multi-cell stage, oblong stage, sphere stage, 50% epiboly stage (50%), 80% epiboly stage (80%), bud stage, 5 somite stage (5s), 18 somite stage (18s), and from 24 hours post fertilization (24 h) were used for qRT-PCR (A) and semi quantitative PCR (B). Ne: negative control. (C) Maternal and uniform distributions of eaf1 and eaf2 in embryos at 4 cell stage, 8-cell stage and multi-cell stage were revealed by WISH. (D) Spatial distribution of eaf1 or eaf2 in embryos at gastrula stage (D1, D2) and at bud stage (D3, D4), and more eaf1 or eaf2 was distributed in the dorsal region of embryos. C1, C3, C4, C5, C7, C8, D3, D4, lateral view, anterior to the up; C2, C6, D1, D2, animal view, dorsal to the right for D1 and D2. |
|
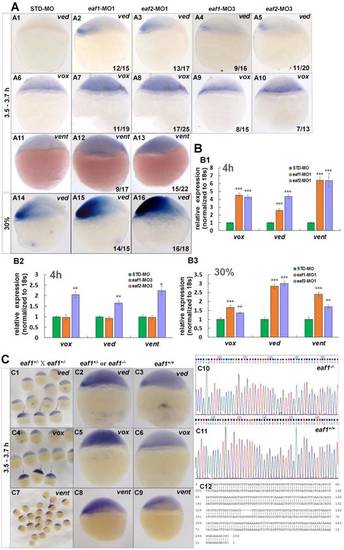
Eaf1 and eaf2 inhibited the expression of early ventral vent family genes at blastula stage. (A) Initial expression of ved (A1-A3), vox (A6-A8) and vent (A11-A13) was increased obviously in embryos injected with eaf-ATG-MO [eaf1-MO1 (8 ng/embryo) and eaf2-MO1 (8 ng/embryo)] or with eaf-splicing-MO (A4, A5, A9, A10) [eaf1-MO3 (8 ng/embryo) and eaf2-MO3 (8 ng/embryo)] at 3.5-3.7 h, and increased expression of ved was also observed in eaf1/2 ATG morphants at 30% epiboly stage (A14-A16). (B) The significantly increased expression of the ventral genes in ATG morphants at 4 h (B1) and at 30% epiboly stage (B3) was revealed by qRT-PCR, and their slightly increased expressionin eaf2 splice morphants but not in eaf1 splice morphants was revealed by qRT-PCR (B2). (C) Expression of ventral ved (C1-C3), vox (C4-C6), and vent (C7-C9) in embryos from in-crossed eaf1 F1 heterozygous mutants, and black arrows indicated the embryos with increased expression of ved (C1), vox (C4), and vent (C7). The genotypic results for embryos with increased expression (C2, C5, C8) or normal expression (C3, C6, C9) were indicted in C10, C11, and C12 respectively. eaf1-MO1: eaf1 ATG morpholino; eaf2-MO1: eaf2 ATG morpholino; eaf1-MO3: eaf1 splicing morpholino; eaf2-MO3: eaf2 splicing morpholino. A1-A16, C1-C9, lateral view, dorsal to the right. |
|
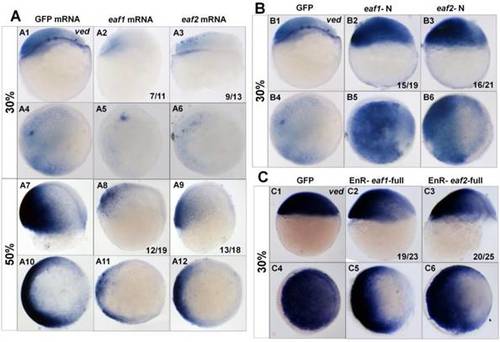
Expression of ved in embryos with ectopic expressions of different forms of eaf1 or eaf2. (A1-A12) Expression of ved was obviously reduced in embryos with ectopic expression of eaf1 or eaf2 at 30% epiboly stage (A1-A6) and at 50% epiboly stage (A7-A12). (B1-B6) Ved expression was obviously increased in embryos injected with eaf1/2-N, dominant forms of eaf1/2. (C1-C6) Expression of ved was obviously reduced in embryos injected with EnR-eaf1/2-full. A1-A3, A7-A9, B1-B3, C1-C3, lateral view, dorsal to the right; A4-A6, A10-A12, B4-B6, C4-C6, animal view, dorsal to the right. |
|
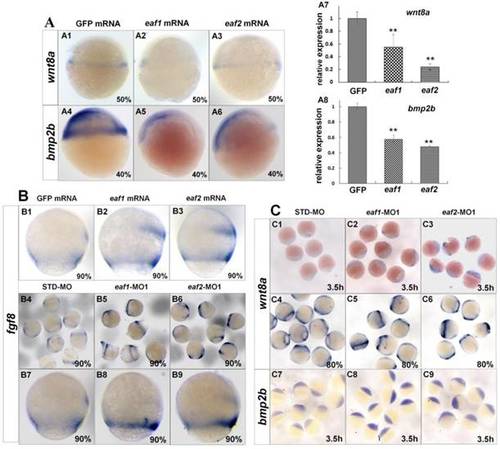
Expression of ligands wnt8a, bmp2b, and fgf8 in eaf1 or eaf2 gain-of-function or loss-of-function embryos. (A) wnt8a (A1-A3, A7) and bmp2b (A4-A6, A8) exhibited reduced expression in eaf1 or eaf2 gain-of-function embryos as detected by in situ and qRT-PCR. (B) Expression of fgf8 was increased in both eaf1/2 gain-of-function (B1-B3) and loss-of-function embryos (B4-B9). (C) Expression of wnt8a (C1-C6) and bmp2b (C7-C9) was increased in eaf1/2 gain-of-function embryos. A1-A6, B1-B9, C1-C9, lateral view. |
|
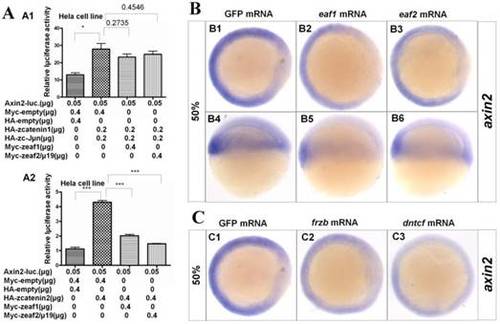
Eaf1 and eaf2 obviously suppressed axin2 expression. (A) Eaf1 and eaf2 suppressed axin2 promoter activity induced by β-catenin2 (A2) but not by β-catenin1 (A1). (B) Expression of axin2 was obviously abolished in the dorsal domain of eaf1/2 gain-of-function embryos. (C) Expression of axin2 was obviously abolished in both dorsal and ventral regions of embryos injected with Wnt antagonist frzb or dntcf. B1-B3, C1-C3, animal view, dorsal to the right; B4-B6, lateral view, dorsal to the right. |
|
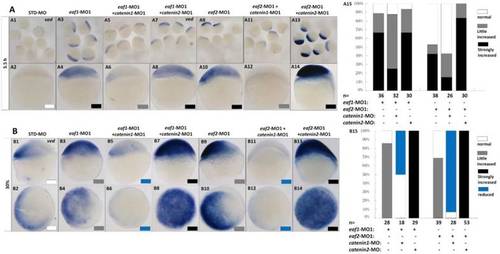
Eaf1 or eaf2 regulated expression of ventral ved gene via Wnt/β-catenin1 signaling. (A, B) β-catenin1-MO (8 ng/embryo) rather than β-catenin2-MO (8 ng/embryo) could recover enhanced ved expression in eaf1 or eaf2 morphants to normal at both oblong stage (A) and 30% epiboly stage (B). Embryos were injected with STD-MO (A1, A2, B1, B2), eaf1-MO1 (A3, A4, B3, B4) or eaf2-MO1 (A9, A10, B9, B10). Embryos were injected with either a combination of eaf1-MO1 and β-catenin1-MO (A5, A6, B5, B6), eaf1-MO1 and β-catenin2-MO (A7, A8, B7, B8), eaf2-MO1 and β-catenin1-MO (A11, A12, B11, B12), or eaf2-MO1 and β-catenin2-MO (A13, A14, B13, B14). (A15, B15) The percentage of embryos exhibiting different expression level of ved was scored at the sphere stage (A15) and at 30% epiboly stage (B15). White box, normal; gray box, slightly increased; black box, strongly increased; blue box, reduced. All of the injections, including MO alone or eaf-MO combined with catenin-MO, were performed using the same batch of embryos produced by a select number of zebrafish to eliminate error caused by embryo variation. A1-A14, B1, B3, B5, B7, B9, B11, B13, lateral view, dorsal to the right; B2, B4, B6, B8, B10, B12, B14, animal view, dorsal to the right. |
|
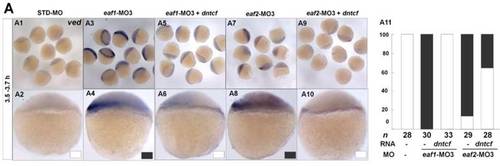
dntcf could effectively rescue enhanced ved expression to normal level in eaf1 or eaf2 morphants. White box, normal; black box, increased. Embryos were injected with STD-MO (A1, A2), eaf1-MO3 (A3, A4) or eaf2-MO3 (A7, A8). Embryos were injected with eaf1-MO3 and dntcf mRNA (10 pg) (A5, A6) or with eaf2-MO3 and dntcf mRNA (10 pg) (A9, A10). All of the injections, including MO alone or MO combined with mRNA, were performed using the same batch of embryos produced by a select number of zebrafish to eliminate error caused by embryo variation. A1-A10, lateral view, dorsal to the right. |