- Title
-
Size-reduced embryos reveal a gradient scaling based mechanism for zebrafish somite formation
- Authors
- Ishimatsu, K., Hiscock, T.W., Collins, Z.M., Sari, D.W.K., Lischer, K., Richmond, D.L., Bessho, Y., Matsui, T., Megason, S.G.
- Source
- Full text @ Development
|
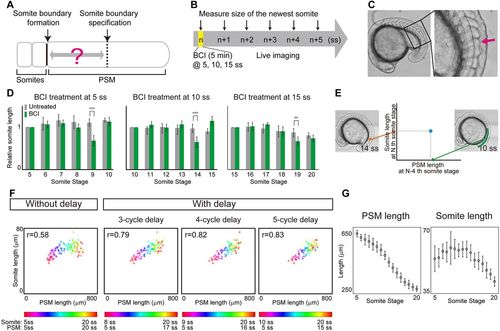
Somite scaling over time with time delay. (A) Schematic of time delay between somite boundary specification and somite boundary formation. (B) Schematic of the BCI experiment. Embryos were treated with BCI for 5 min and then subjected to live imaging in egg water without BCI. The BCI treatment was carried out at three different somite stages (5, 10, 15 ss), in case the delay time varies over time. (C) BCI-treated embryos form smaller somites (arrow). (D) Relative AP length of somites, normalized by the somite length of control embryos at the somite stage at which BCI treatment was carried out. At each somite stage, the smaller somite was formed four cycles after BCI treatment. Error bars denote s.d. **P<0.01; ***P<0.001 (n=5 for each condition). (E) Comparison of PSM length and somite length was made using PSM length at N−4 ss (e.g. 10 ss) and somite length at N ss (e.g. 14 ss), using live imaging data. (F) Somite size versus PSM size at different somite stages (indicated by the color scale) with and without time delay of three, four and five cycles (n=7). (G) Size dynamics of PSM and somites. Note that the peaks appear at different somite stages. n=7 each. Error bars denote s.d. |
|
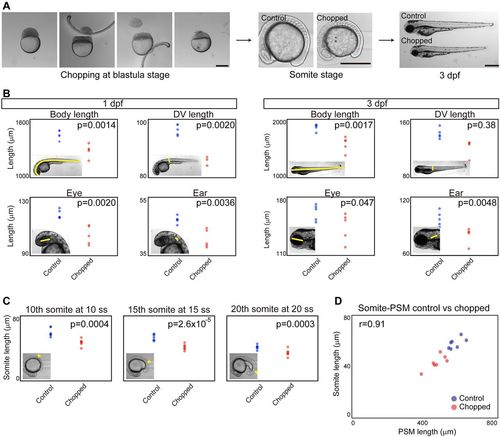
Somite scaling between individuals of different sizes. (A) Size reduction technique. Scale bars: 500 μm. dpf, days post-fertilization. (B) Comparison of body and organ sizes between control and chopped embryos. Insets illustrate the region measured. n=5 each. (C) Comparison of somite size between control and chopped embryos. Somite of interest indicated by arrowhead in insets. n=5 each. (D) Somite size versus PSM size in control and chopped embryos. n=7 each. |
|
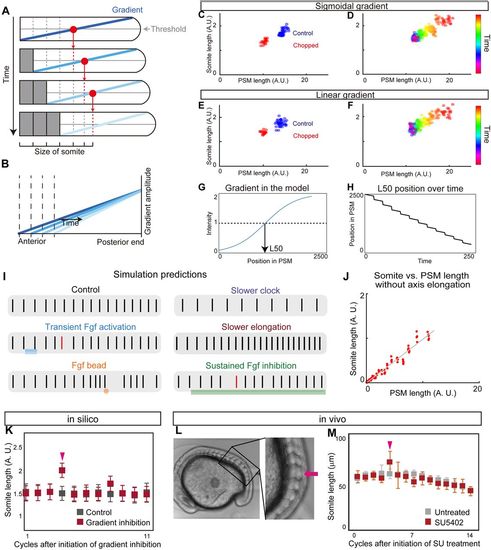
Clock and scaled gradient model. (A) Schematic of the clock and scaled gradient model. The position of a future somite boundary is set by a scaled gradient once per clock cycle (each row) and the boundary appears after a delay. (B) Superimposition of the gradients from each time point shown in A. (C,D) Simulation results using a sigmoidal gradient. (E,F) Simulation results using a linear gradient. (C,E) Simulation results of control and chopped embryos. (D,F) Simulation results of a single embryo over time. (G,H) Stepwise regression of the gradient in clock and scaled gradient model. (G) L50 in the model was determined similarly to Fig. 3P. (H) Clock and scaled gradient model predicts stepwise regression of L50 position. (I) Simulation results for perturbation experiments for local or global inhibition/activation of Fgf, slower clock and slower axis elongation. Black vertical lines represent simulated somite boundary positions. (J) Somite size versus PSM length shows perfect scaling in silico when axial elongation speed is zero, mimicking the results from the in vitro mPSM system (Lauschke et al., 2013). (K) Simulation results of long-term suppression of a gradient in the clock and scaled gradient model. Error bars denote s.d. (L,M) Treatment with a low concentration of SU5402 (16 μM) results in one or two larger somite(s) (arrow) (n=7 for both SU5402 and untreated groups). Error bars denote s.d. |
|
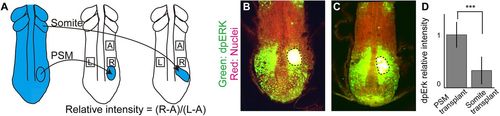
New somites inhibit Fgf activity. (A) Schematic of somite transplantation. (B,C) dpErk immunostaining. Dashed line encircles transplanted tissue. (D) Comparison of relative intensities between PSM-transplanted samples (n=9) and somite-transplanted samples (n=9). Error bars denote s.d. ***P<0.001. |
|
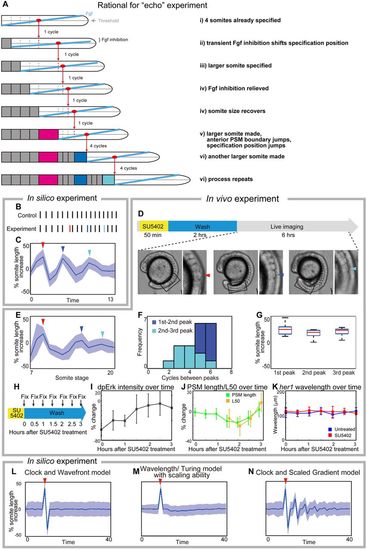
Experimental validation of the clock and scaled gradient model. (A) Schematic of the outcome of the clock and scaled gradient model, following induction of one larger somite. The induced larger somite is magenta, and the larger somites as a result of system response are blue and cyan. (B,C) Simulation results without (B) and with (C) noise for somite size (red line in B, red arrowhead in C). Blue line in B and blue arrowhead in C show the second, and cyan line in B and cyan arrowhead in C show the third large somite. (D) Schematic of the in vivo experiment, and an embryo with larger somites at different time points. Boxed areas are shown at higher magnification to the right with arrowheads indicating the larger somite. (E) Time course of percentage increase in somite length of SU5402-treated embryos, compared with those in control embryos (n=12). (F) Frequency distribution of somite cycles between the peaks. (G) Percentage increase in somite size in SU5402-treated embryos at the peaks detected in each embryo, compared with control embryos at the corresponding somite stage. In both C and E, blue lines and blue shades indicate the average somite size and the variance of one standard deviation, respectively. For C-E, red, blue and cyan arrowheads show the first, second and third larger somites, respectively. (H-K) Examination of Erk activity (n=6 for each time point) and her1 wavelength (n=4-8 for each time point) after transient SU5402 treatment. Error bars denote s.d. (H) Schematic of the experiment. After fixation, the samples were subjected to dpErk immunostaining and her1 in situ hybridization. (I) Time course of percentage change in dpErk maximum intensity in SU5402-treated embryos, compared with that in control embryos. (J) Time course percentage change in PSM size and L50 position in SU5402-treated embryos, compared with those in control embryos. Note that L50 analysis begins 1.5 h after SU5402 treatment when dpErk intensity has recovered (see Fig. 6I) because it cannot be defined earlier. (K) Time course analysis of her1 wavelength of untreated embryos and SU5402-treated embryos. We found no significant difference (significance threshold P<0.05) at any time point. (L-N) Simulation results for percentage increase of somite size over time, based on different models. After induction of one larger somite (arrowheads), the clock and wavefront model (when wavefront speed is associated with axis elongation only) predicts one smaller somite (L), the wavelength/Turing model (Cotterell et al., 2015) predicts smaller somites and the somite size eventually comes back to normal (M). Only the clock and scaled gradient model predicts the ‘echo effect’ that somite size dynamics oscillate repeatedly every four cycles (N). |
|
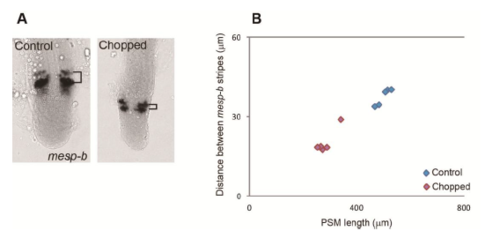
Scaling of mesp-b stripe. (A) in situ hybridization samples of mesp-b. (B) PSM length vs distance between mesp-b stripes. |
|
Long-term SU5402 treatment under constant dark condition The embryos were treated with SU5402 at low concentration (16M) for 4 hrs with the light completely blocked. One or two larger somites were formed (magenta arrow in the right panel) several cycles after initiation of the treatment (10 out of 11). |