- Title
-
Multiple zebrafish atoh1 genes specify a diversity of neuronal types in the zebrafish cerebellum
- Authors
- Kidwell, C.U., Su, C.Y., Hibi, M., Moens, C.B.
- Source
- Full text @ Dev. Biol.
|
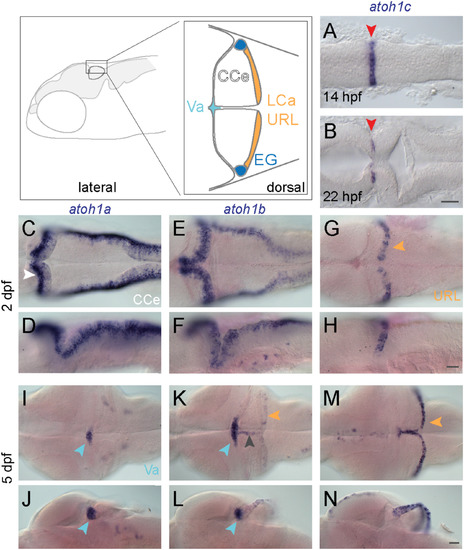
atoh1 gene expression in the developing zebrafish cerebellum. Schematic depicts regions of the zebrafish cerebellum in lateral (left panel) and dorsal (right panel) views at 5 dpf. Throughout the manuscript, white arrowheads indicate the CCe, light blue arrowheads indicate the Va, orange arrowheads indicate the URL (at early stages) or the LCa (at later stages) and dark blue arrowheads indicate the EG. Red arrowheads indicate the position of the MHB. A-N: RNA in situ hybridization in wild-type embryos with atoh1a (left column), atoh1b (middle column), and atoh1c (right column) at 14 hpf (A) 22 hpf (B), 2 dpf (C-H), and 5 dpf (I-N). Dorsal (A,B,C,E,G,I,K,M) or lateral (D,F,H,J,L,N) views are shown with anterior to the left. Gray arrowhead indicates midline of CCe. CCe, corpus cerebelli; EG, eminentia granularis; LCa, lobus caudalis cerebelli; URL, upper rhombic lip; Va, valvula cerebelli; Scale bars: 50 μM. EXPRESSION / LABELING:
|
|
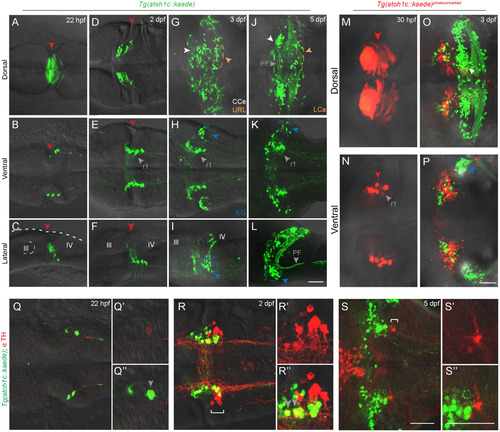
atoh1c-expressing progenitors give rise to to ventral r1 and cerebellar granule neurons. A-L: Tg(atoh1c::kaede) transgene expression in fixed embryos stained with anti-Kaede antibody in dorsal (A,D,G,J), ventral (B,E,H,K) and lateral (C,F,I,L) views at 22 hpf (A-C), 2 dpf (D-F), 3 dpf (G-I) and 5 dpf (J-L). Arrowheads follow the color code described in Fig. 1 legend or are labeled as follows: PF: parallel fibers, r1: MHB-derived neurons in ventral r1. M-P: live imaging after photoconversion of Kaede (green to red) of MHB atoh1c+ cells at 22 hpf confirms that this Tg(atoh1c::kaede)+ progenitor domain gives rise to ventral r1 neurons. Dorsal (M-O) and ventral focal planes (N-P). Q-S: Tg(atoh1c::kaede)+ cells (green) transiently express TH (red; gray arrowheads) from 22 hpf (Q) to 2 dpf (R) and lie adjacent to the LC (indicated by white bracket). All images oriented with anterior to the left at time points as indicated. Scale bars: 50 μM. EXPRESSION / LABELING:
|
|
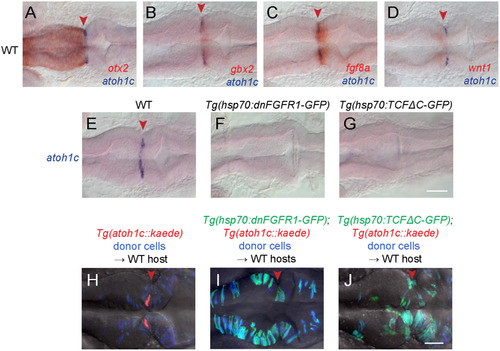
Specification of the atoh1c MHB domain. A-D: Double RNA in situs for atoh1c (blue) and otx2 (A), gbx2 (B), fgf8 (C), and wnt1 (D) (red) show that the atoh1c expression in a few cells immediately posterior to the MHB. E-G: atoh1c expression is absent when Fgf (F) or Wnt (G) signaling is blocked with heat-inducible dominant-negative transgenes activated at 10 hpf. H-J: atoh1c expression requires cell-autonomous Fgf and Wnt signaling. Live imaging of chimeras with donor-derived cells (blue) expressing dn-FGFR1 (I, green) or dn-TCFΔC (J, green) that are unable to express Tg(atoh1c::kaede) (red in the control chimera in H) even if they lie at the MHB (red arrowheads). All embryos are at 22 hpf and are shown in dorsal views with anterior to left. Scale bars: 50 µM. EXPRESSION / LABELING:
PHENOTYPE:
|
|
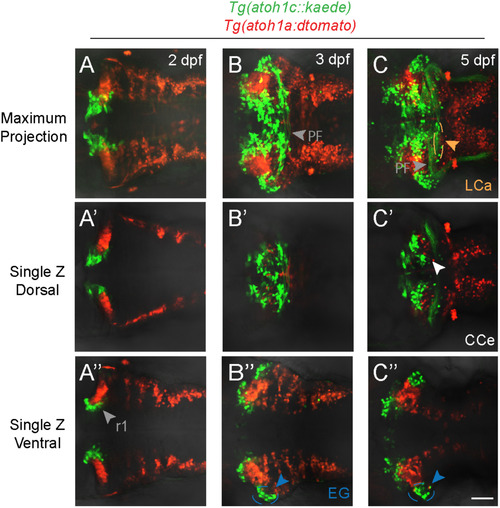
atoh1a- and atoh1c-derived neurons specify distinct progenitor pools. Live imaging of wild-type embryos with Tg(atoh1c::kaede) (green) and Tg(atoh1a:dtomato) (red) transgenes indicate atoh1a and atoh1c-derived neurons are distinct cerebellar populations. Maximum projections of a z-stack (A-C) and single z-slices at dorsal (A′-C′) or ventral (A”-C”) focal planes with anterior to the left at time points as indicated. Scale bar: 50μM. |
|
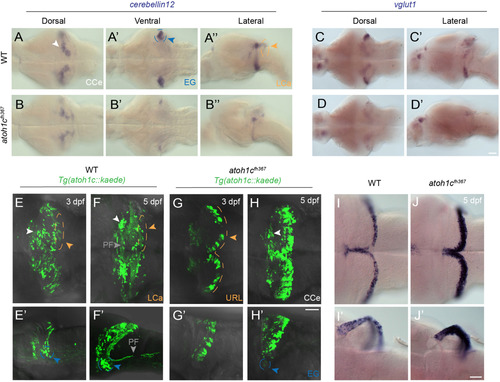
atoh1c is required for cerebellar granule cell fate. A-D: Reduction in the number of mature GCs in atoh1c mutant as indicated by decreased staining for cerebellin12 (B) and vglut1 (D), at 5 dpf. E-H: The majority of mutant cells expressing Tg(atoh1c::kaede) remain as a non-migrated population in the URL at 3 dpf (G, G′ compare to wild-type in E, E′) and 5 dpf (H, H′ compare to wild-type in F, F′). I-J: Upregulation of atoh1c mRNA in URL at 5 dpf in atoh1c mutant embryos (J, J′ compare to I, I′). Dorsal (A,B,C,D,E,F,G,H,I,J), ventral (A′,B′) and lateral (A”,B”,C′,D′,E′,F′,G′, H′,I′,J′) views are shown with anterior to the left. Scale bars: 50 μM. EXPRESSION / LABELING:
PHENOTYPE:
|
|
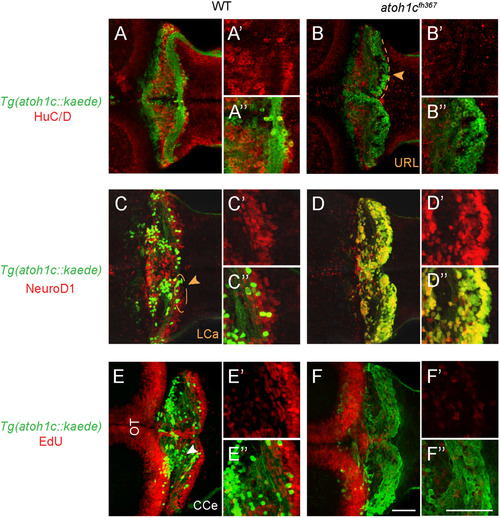
atoh1cfh367 cerebellar cells accumulate as post-mitotic but undifferentiated granule cell progenitors. A,B: The majority of Tg(atoh1c::kaede)+ cells (green) in the atoh1c mutant are located within the URL and do not express HuC/D (red) at 5 dpf (B) indicating that they are not post-mitotic neurons. C,D: atoh1cfh367 cells (green) in the URL are positive for NeuroD1 (red, D). E,F: atoh1cfh367 cells (green) in the URL do not incorporate EdU (red, F). Strong EdU incorporation anterior to the cerebellum in both wild-type and mutant is in the optic tectum (OT). Dorsal views with anterior to the left. Scale bars: 50 μM. |
|
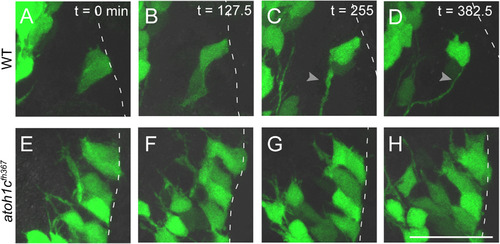
Atoh1c is required for release of granule neuron progenitors from URL. Still images from confocal timelapses taken at 3 dpf. Dotted line indicates the URL where the apical surfaces of GC progenitors attach. In wild-type, Tg(atoh1c::kaede)+ progenitors detach from the URL over a six-hour interval (A-D) while in atoh1c mutants, Tg(atoh1c::kaede)+ cells remain attached at the URL (E-H). Time points indicated in upper right corner in minutes. Scale bar: 25 µM. EXPRESSION / LABELING:
PHENOTYPE:
|
|
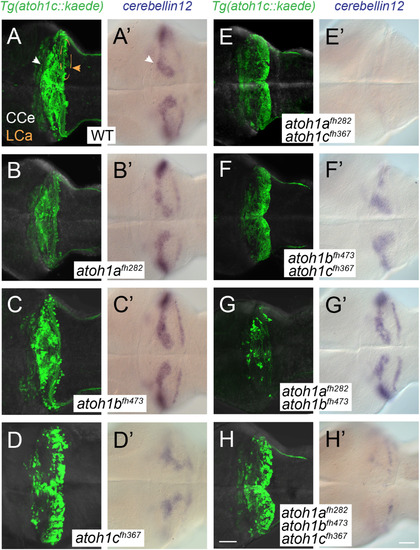
atoh1a, atoh1b, and atoh1c have non-overlapping roles in cerebellar development. Images of 5 dpf wild-type (A), single (B-D), and compound (E-H) atoh1 mutants with the Tg(atoh1c:kaede) (left column) or cbln12 RNA in situ hybridization (right column). Dorsal views with anterior to the left. Scale bars: 50 μM. EXPRESSION / LABELING:
PHENOTYPE:
|
|
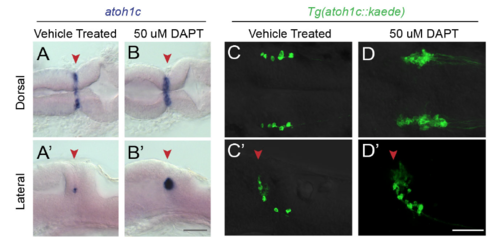
Notch-mediated lateral inhibition of atoh1c MHB domain. Pharmacological inhibition of Notch signaling with g-secretase inhibitor DAPT blocks restriction of atoh1c expression domain (B) resulting in an overproduction of Tg(atoh1c::kaede)+ neurons (D) compared to stage-matched vehicle treated control embryos (A,C). 22 hpf embryos (A,B) treated for 18 hours or 30 hpf embryos (C,D) treated for 26 hours in orientations as indicated.Red arrowheads indicate location of MHB. Scale bars: 50 uM. |
Reprinted from Developmental Biology, 438(1), Kidwell, C.U., Su, C.Y., Hibi, M., Moens, C.B., Multiple zebrafish atoh1 genes specify a diversity of neuronal types in the zebrafish cerebellum, 44-56, Copyright (2018) with permission from Elsevier. Full text @ Dev. Biol.