- Title
-
Comprehensive analysis of target genes in zebrafish embryos reveals gbx2 involvement in neurogenesis
- Authors
- Nakayama, Y., Inomata, C., Yuikawa, T., Tsuda, S., Yamasu, K.
- Source
- Full text @ Dev. Biol.

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Altered expression of brain-forming genes due to the induction of gbx2. Embryos obtained by mating Tg(hsp70l:gbx2) and wild-type fish were subjected to heat shock at the bud stage and analyzed by WISH at 2 hph. The genotypes of the stained embryos, shown above (a–f, sibling; a'–f', hsp-gbx+), were confirmed by PCR. Images show dorsal views, oriented with anterior to the top. Scale bars, 200 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
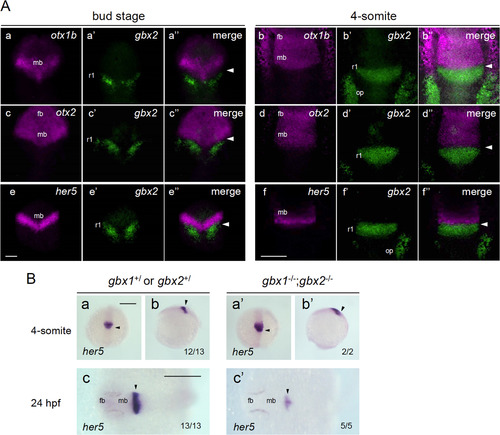
Close relationship between gbx2 and regionalization genes. A. Closely associated expression of gbx2 and its target genes around the MHB. The expression of gbx2 and three candidate target genes (otx1b, otx2, and her5) was detected by fluorescence in situ hybridization (FISH) in wild-type embryos at the bud stage (left three columns) or 4-somite stage (right three columns). Dorsal views with anterior to the top. The expression of the candidate target genes (magenta), gbx2 (green), and their merged views are shown for each gene and stage. Scale bars, 100 µm. B. The expression of her5 was examined in embryos obtained by crossing double gbx heterozygotes (gbx1+/–;gbx2+/–) at the 4-somite stage (a, b, a', b') and 24 hpf (c, c'). Right three panels (a'–c') show her5 expression in double homozygotes and the left three (a–c) show expression in the rest of the offspring. The genotypes of the stained embryos were confirmed by PCR. Images show dorsal views with anterior to the top (a, a') and left (c, c'), and lateral views with anterior to the top and dorsal to the right (b, b'). fb, forebrain; mb, midbrain; op, otic placode; r1/2/4, rhombomere 1/2/4; arrowhead, midbrain-hindbrain boundary (MHB). Scale bars, 200 µm. PHENOTYPE:
|
|
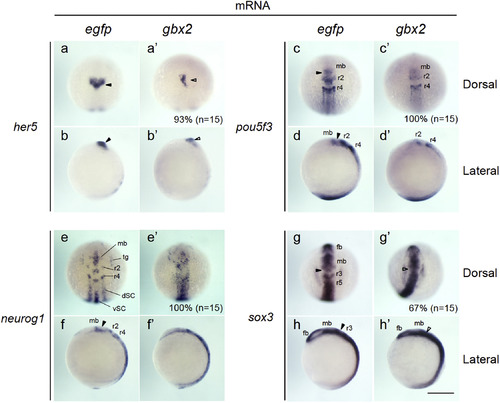
Expression of neurogenesis-related genes in embryos injected with gbx2 mRNA. The expression of genes related to neural development was examined by whole-mount in situ hybridization (WISH) in 3-somite-stage embryos injected with mRNA for egfp (a–h) or gbx2 (a'–h'). (a, c, e, g, a', c', e', g') Dorsal views with anterior to the top. (b, d, f, h, b', d', f', h') Lateral views with anterior to the top and dorsal to the right. Percentages of the embryos showing abnormal expression are shown at the bottom right with total numbers of stained embryos in parentheses. dSC, dorsal spinal cord; fb, forebrain; mb, midbrain; r2–r5, rhombomeres 2–5; tg, trigeminal ganglion; vSC, ventral spinal cord; black arrowheads, normal MHB; open arrowheads, abnormal MHB. Scale bars, 200 µm. |
|
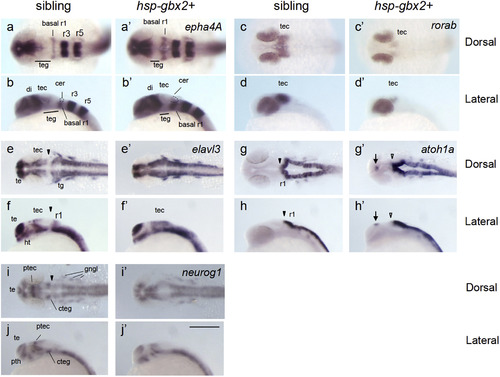
gbx2 promotes neurogenesis in the midbrain-hindbrain region. Embryos obtained by mating Tg(hsp70l:gbx2) and wild-type fish were subjected to heat shock at the bud stage and analyzed at 24 hpf for the expression of neurogenesis genes by WISH. The genotypes of the stained embryos were confirmed by PCR (a–j, sibling; a'–j', hsp-gbx2+). Dorsal views with anterior to the left (a, c, e, g, i, a', c', e', g', i') and lateral views with anterior to the left and dorsal to the top (b, d, f, h, j, b', d', f', h', j') are shown. Arrows, ectopic expression; cer, cerebellum; basal r1, basal region of rhombomere 1; cteg, caudal tegmentum; di, diencephalon; gngl, cranial ganglion; ht, hypothalamus; ptec, pretectum; pth, prethalamus; r1/r3/r5, rhombomere 1/3/5; te, telencephalon; tec, tectum; teg, tegmentum; tg, trigeminal ganglion; black arrowheads, normal MHB; open arrowheads, abnormal MHB. Scale bars, 200 µm. |
|
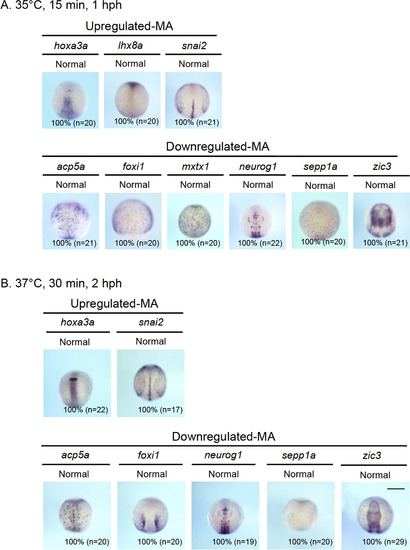
Whole-mount in situ hybridization (WISH) analysis of genes whose expression was shown by the microarray data to be altered by transient gbx2 induction. The expression of the genes shown by the microarray analysis (MA) to be significantly upregulated (Upregulated-MA) or downregulated (Downregulated-MA) was re-examined by WISH in embryos that were exposed to two modes of heat shock (A, 35 °C for 15 min and fixed 1 h post-heat shock (hph); B, 37 °C for 30 min and fixed 2 hph). For the genes shown here, no differences were observed, in contrast to the genes shown in Fig. 3. |
|
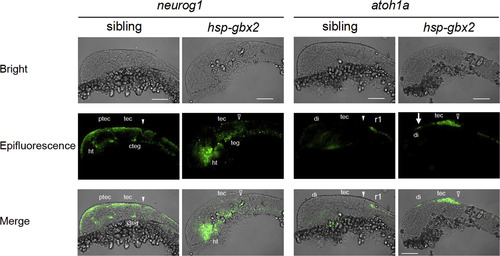
Fluorescent in situ hybridization analysis of the effects of hsp-gbx2 induction on proneural genes. Embryos obtained by mating Tg(hsp70l:gbx2) and wild-type fish were subjected to heat shock at the bud stage and analyzed at 24 hpf for the expression of proneural genes by fluorescent in situ hybridization. The genotypes of the stained embryos were confirmed by PCR. Lateral views with anterior to the left and dorsal to the top are shown. Arrows, ectopic expression in the diencephalon; cteg, caudal tegmentum; di, diencephalon; ht, hypothalamus; ptec, pretectum; r1, rhombomere 1; tec, tectum; teg, tegmentum; solid and open arrowheads, normal and disrupted MHB, respectively. Scale bars, 100 µm. |
|
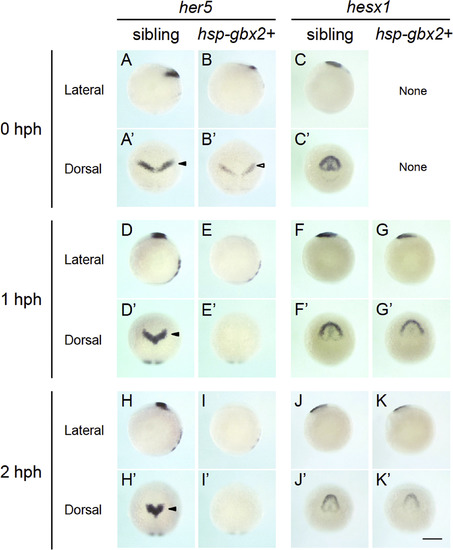
Time course of the expression of genes shown to be downregulated by transient gbx2 induction. The expression of the two brain regionalization genes, shown by the microarray data to be downregulated, were reexamined by WISH at 0, 1, and 2 hph in sibling and hsp-gbx2+ embryos. The data show that her5 is immediately downregulated, whereas hesx1 responded much later, suggesting indirect regulation by gbx2. None, no abnormal expression was observed. Scale bars, 200 µm. |
Reprinted from Developmental Biology, 430(1), Nakayama, Y., Inomata, C., Yuikawa, T., Tsuda, S., Yamasu, K., Comprehensive analysis of target genes in zebrafish embryos reveals gbx2 involvement in neurogenesis, 237-248, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.