- Title
-
Hecate/Grip2a acts to reorganize the cytoskeleton in the symmetry-breaking event of embryonic axis induction
- Authors
- Ge, X., Grotjahn, D., Welch, E., Lyman-Gingerich, J., Holguin, C., Dimitrova, E., Abrams, E.W., Gupta, T., Marlow, F.L., Yabe, T., Adler, A., Mullins, M.C., Pelegri, F.
- Source
- Full text @ PLoS Genet.
|
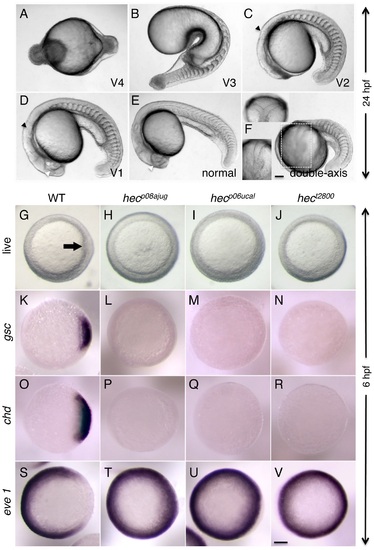
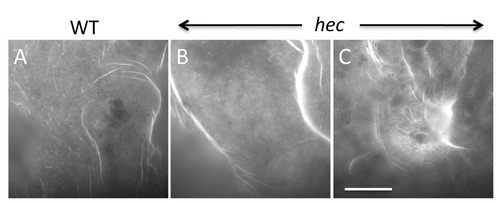
Axis induction defects in embryos from mothers homozygous for three different hecate mutant alleles. A-F) Side views (except as indicated in (F)) of live embryos at 24 hpf showing a range of phenotypes from most severe (A) to normal (E) and double-axis embryos (F). A) Class V4 embryo exhibiting complete radial symmetry and lacking all anterior (e.g. head structures) or dorsalmost (e.g. notochord) structures. B) Class V3 embryo exhibiting a rudimentary axis but lacking all anterior and dorsalmost structures. C) Class V2 embryo lacking anteriormost structures such as the eyes, but showing formation of more posterior head structures such as the otic vesicles (black arrowhead, also indicated in D). D) Class V1 embryo with a relatively complete axis but reduced anterior structures, such as the eyes (white arrowhead, also labeled in E), and lacking a properly formed notochord. E) Normal embryo derived from hec mutant females indistinguishable from wild-type. Note somites in (E) are chevron-shaped, while they are blocky in (B-D) indicative of defects in notochord formation, or encircling the entire embryo in (A). F) A double-axis embryo found in hec mutant clutches exhibiting weak expressivity. Insert in the lower left shows the left axis indicated by the dashed rectangle, out of focus in the main image. Insert in the upper left panel shows a dorsal view, showing the bifurcated axis. Note the lack of defined anterior structures in both axes, as well as the lack of a notochord along the trunk, also reflected by block-shaped somites in this region. Images in A–F are side views, except for upper left insert in (F). G-I) Animal views of live wild-type embryos (G) and embryos from females homozygous for each of the three hec mutant alleles. The dorsal thickening or shield (arrow) is absent in mutant embryos. K-V) In situ hybridization analysis to detect expression of dorsally-expressed genes (gsc, chd) and ventrally-expressed genes (eve 1). The expression domains of gsc and cho is reduced in hec mutant embryos, while the expression domain of eve is expanded in these embryos. (K-V) are animal view of embryos, dorsal to the right when identifiable, at the shield stage (6 hpf). Magnification bars in (F) and (V) correspond to 100 µm for panels sets (A-F) and (G-V), respectively. Dorsal view insert in panel (F, upper left) has been reduced to 75% size. |
|
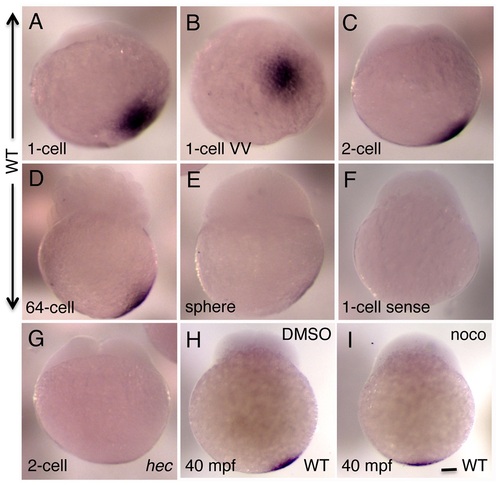
Whole mount in situ hybridization analysis of the expression of zebrafish grip2a mRNA in early embryos. A-E) grip2a mRNA is localized in a slightly off-center position at the vegetal pole of early embryos, and remains in that position until the sphere stage when it becomes undetectable. F) Control sense probe. G) The domain of grip2a mRNA localization in hec mutants is reduced in intensity and appears to lack an off-center shift. H,I) Representative control (DMSO)- and nocodazole-treated embryos showing the lack of off-center shift in nocodazole-treated embryos (see Figure S4 for details). All panels are side views (except for (B), which is a vegetal view of the embryo in (A)) at the following stages: A,B: 1-cell (20 mpf), C,F,G: 2-cell (45 mpf), D: 64-cell (2 hpf), E: sphere (4 hpf). (H,I) are at 40 mpf, which approximately corresponds to the 2-cell stage (C) in untreated embryos. Magnification bar in (I) corresponds to 100 µm for all panels. |
|
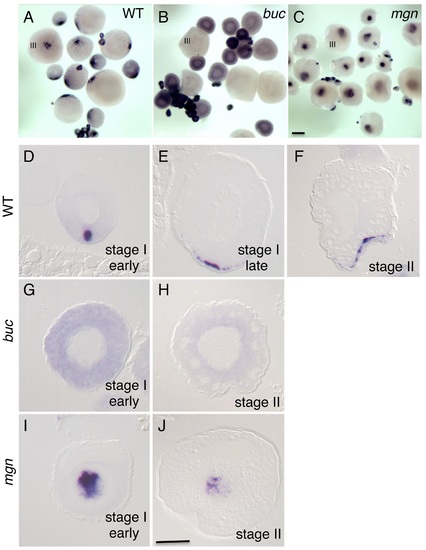
Localization of grip2a mRNA in wild-type and mutant oocytes. A-C) Whole mount in situ hybridization of dissected ovaries from wild-type (A), bucky ball (buc, B) and magellan (mgn, C) mutant females. In (A-C) oocytes at stage III of development are indicated. Smaller oocytes are at stages I and II, which are difficult to differentiate in whole mounts at this magnification. The grip2a mRNA localization domain is observed in an asymmetric cortical position in wild-type oocytes (A) but is unlocalized and diffuse in buc oocytes (B) and internally-located in mgn mutants (C). D-J) Sections of wild-type and mutant oocytes at the indicated stages after labeling to detect grip2a mRNA. Stages are as indicated in the panel and were determined by size and oocyte morphology according to [101]. D–F) Wild-type oocytes showing localization to the presumptive Balbiani Body (D) and subsequent localization to a cortical domain of the oocyte corresponding to the presumptive vegetal pole (E-F). G,H) buc mutant oocytes lack the Balbiani body [39], [43] and the grip2a mRNA subcellular localization domain in stage I and II oocytes. I) mgn mutant stage I oocytes exhibit an enlarged Balbiani body [40] and displayed an enlarged grip2a mRNA localization domain. J) Stage II mgn mutant oocytes fail to localize transcripts to the vegetal pole which instead persist in an internal domain [40], as observed also for grip2a mRNA. Number of oocytes examined were as follows: wild-type: early stage I: 13, stage II: 12; buc: early stage I: 35, stage II: 26; mgn: early stage I: 23, stage II: 18. Magnification bar in (C) corresponds to 250 µm for panels (A-C), and in (J) to 50 µm for panels (D–J). |
|
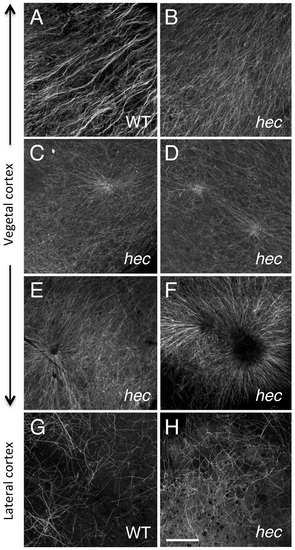
Microtubule reorganization at the vegetal cortex is affected in hecate mutants. (A-F) Cortical microtubule network at the vegetal pole in wild-type (A) and hec mutant (B-F) embryos at 20 mpf. Microtubules appear oriented in the same direction and bundled in wild-type embryos (A). The extent of co-orientation and bundling is greatly reduced in hec mutant embryos (B), where microtubules form multiple aster-like structures which can have a well focused-center (C,D) or can exhibit a central microtubule-free zone (E,F) and often overlap (F) or interdigitate (D). The relatively unbundled microtubule arrangement shown in (B) also corresponds to a sector of a large aster-like structure emanating from a not shown central core. Up to 6 aster-like structures were observed in the vegetal cortex of a single embryo. G,H) Cortex in mediolateral regions shows a loose and apparently random network of microtubules which appears similar in both wild-type (G) and hec mutant (H) embryos (n = 8 for wild-type and mutants). All images are z-axis projections of confocal image stages. The phenotype was fully (100%) penetrant according to the two main categories (wild-type, aligned and bundled microtubules; mutant, radialy oriented and unbundled microtubules, with 10 wt and 25 hec mutant embryos imaged. Magnification bar in (H) corresponds to 40 µm for all panels. PHENOTYPE:
|
|
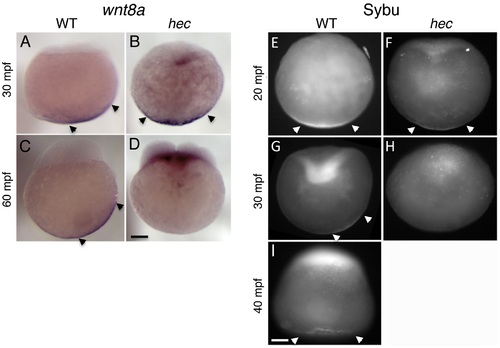
Defects in the vegetal localization of wnt8a mRNA and Sybu protein. (A-D) Off-center shift of wnt8a mRNA is affected in hec mutants. Whole mount in situ hybridization of wild-type embryos (A,C) and hec mutant embryos (B,D) at the 1- (A,B, 30 mpf) and 4- (C,D, 60 mpf) cell stages. Images show representative embryos. A majority of wild-type embryos showed a clear off-center shift (85%, n = 27 at 30 mpf and 74%, n = 47 at 60 mpf). A majority of hec mutant embryos showed vegetal localization without a shift at 30 mpf (79%, n = 33, remaining embryos show no localization) and absence of localization at 60 mpf (89%, n = 38, remaining embryos show reduced vegetal localization without a shift). The apparent label at the base of the blastodisc is observed in a majority of mutant embryos (71%, n = 38) but not in wild-type (C) or control embryos labeled with other probes (not shown) and may reflect remaining wnt8a mRNA that has lost anchoring at the vegetal pole and has moved animally through the action of axial streamers [83]. E–I) Localization of Sybu protein is affected in hec mutants. Whole mount immunofluorescence to detect Sybu protein of untreated wild-type (E,G) and hec mutant (F,H,) embryos and nocodazole-treated wild-type embryos (I) at the indicated stages. In wild-type embryos, an off-center shift in Sybu protein can be observed starting at 30 mpf (G). In hec mutants, Sybu protein becomes undetectable levels by this same time point (H). Patterns of localization of Sybu protein at 10 mpf and 20 mpf time points (combined n: 32 WT, 19 mutant for 10–20 mpf), and 30 mpf and 40 mpf time points were similar and have been combined. 59% (n = 32) of wild-type and 63% (n = 19) of hec mutant embryos showed centered vegetal localization during 10–20 mpf. At 30–40 mpf, the percent of embryos that showed vegetal localization, now with an off-center shift, was reduced to 25% (n = 28) in wild-type, and 0% (n = 25) of hec mutants showed any localization at these time points. Treatment of wild-type embryos with nocodazole inhibits the shift but does not result in delocalization from the vegetal cortex (I, embryo at 40 mpf), as previously shown [6]. Magnification bars in (D) and (I) correspond to 100 µm for panels sets (A-D) and (E-I), respectively. |
|
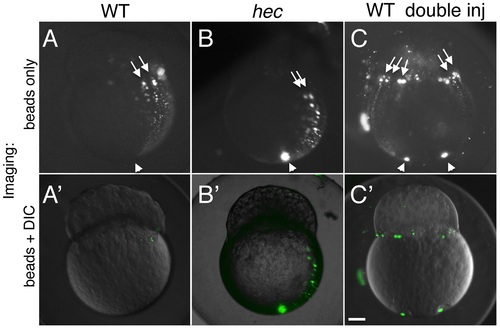
Long-range animally-directed transport is not affected in hecate mutants. (A-C) Paths of injected 0.2 um fluorescent beads after injection into the vegetal pole with a single (A,B) or double (C) injection in wild-type (A,C) or hec mutant (B) embryos. (A′-C′) show merged imaged including the fluorescent channel (shown in A-C) and corresponding DIC optics at low intensity. The extent and frequency of bead transport appeared similar in wild-type and mutant embryos (A,B, see text). Injections into two opposite sides of the vegetal pole results in multiple animally-directed paths, indicating that the entirety of the mediolateral cortex is competent for bead movement. Arrowheads and arrows in (A-C) indicate site of injection in the vegetal region and animally-directed paths along mediolateral regions, respectively. Magnification bar in (C′) corresponds to 100 µm for all panels. PHENOTYPE:
|
|
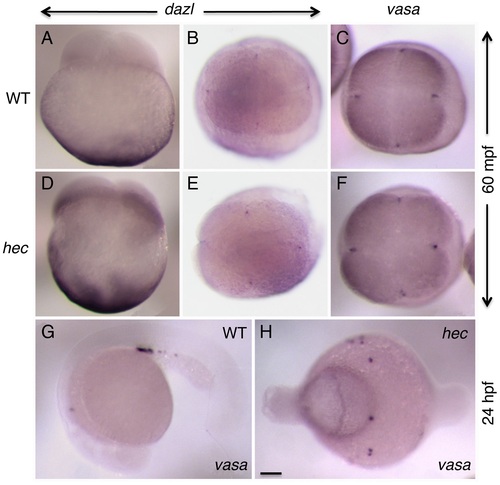
Germ plasm recruitment and PCG determination appears unaffected in hecate mutants. (A-F) Whole mount in situ hybridization of 4-cell stage (60 mpf) embryos to label the germ line-specific genes dazl1 (A,B,D,E), and vasa (C,F) in wild-type (A-C) and hec mutant (D–F) embryos. Early localization of dazl mRNA to the vegetal pole (A,D, side views), and dazl mRNA and vasa mRNA to the germ plasm as it becomes recruited to the furrows of the first and second cleavage cycles (B,C,E,F, animal views). Localization of dazl domains of recruitment at the furrow is not detected in the side views as the levels of mRNA in these domains are relatively low and the focal plane is not optimal for their visualization. G,H) PGC determination, as determined by vasa expression in 24-hour embryos (G), appears unaffected in hec mutants (H), although the PGCs in mutant embryos do not become clustered as in wild-type due to the aberrant cell specification in these mutants. Quantification of the number of vasa-expressing cells is presented in Figure S9. Magnification bar in (H) corresponds to 100 µm for all panels. |
|
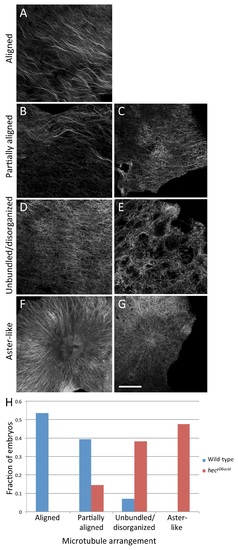
Distribution of microtubule organization phenotypes at the vegetal cortex of wild-type and hecate mutant embryos. A-G) Examples of various vegetal cortex microtubule arrangements at 20 mpf: normally aligned (A), partially aligned (B, C), unbundled and lacking organization (D, E) and exhibiting aster-like structures (F, G). (A) is from a wild-type embryo and (B-G) from hec mutants. Magnification bar in (G) corresponds to 40 µm for panels (A-G). H) Distribution of phenotypes. Wild-type embryos exhibit highly aligned microtubule network while hec mutants show disorganization, lack of bundling and aster-like structures. The two distributions are significantly different (Fisher′s Exact Test, p-value = 5.335e-09). |
|
F-actin cortex at the vegetal pole is similar in wild-type and hec mutant embryos. At 20 mpf, both wild-type (A) and mutant (B) eggs show F-actin rich folds and villi-like structures, which may correspond to previously described microplicae [45]. Number of embryos tested: 18 wt (from a pool of four females) and 24 mutants (from two different mutant females). A fraction (21%, n = 24) of hec mutant embryos show radial F-actin enrichments (C), correlating with aster-like microtubule structures in these embryos (Figure S5). Magnification bar in (C) corresponds to 40 µm in all panels. |