- Title
-
Kinesin-1 interacts with Bucky ball to form germ cells and is required to pattern the zebrafish body axis
- Authors
- Campbell, P.D., Heim, A.E., Smith, M.Z., Marlow, F.L.
- Source
- Full text @ Development
|
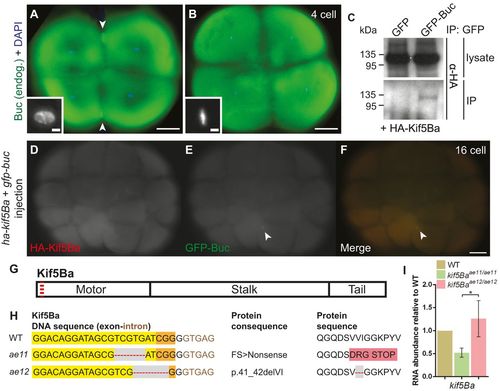
Endogenous Buc localizes to the germ plasm of embryos and binds Kinesin-1. (A,B) Endogenous Buc localizes to distal cleavage furrows of 4-cell embryos when chromosomes are decondensed (A), but not during metaphase (B) or anaphase. Insets show DAPI. Arrowheads indicate Buc accumulation. Scale bars: 100µm for main images, 10µm for insets. See text for quantification. (C) GFP-Buc, but not GFP, co-immunoprecipitates HA-Kif5Ba in HEK293T cells. (D-F) Overexpressed GFP-Buc localizes to cleavage furrows and overexpressed HA-Kif5Ba is expressed throughout blastomeres, potentially allowing them to interact in vivo. Arrowheads indicate Buc accumulation. (G) Schematic protein structure of Kif5Ba, illustrating motor, stalk and tail domains. Red dashed line indicates CRISPR target site. (H) DNA and predicted protein sequences of WT and mutant kif5Ba alleles (ae11 and ae12). Yellow indicates protospacer, orange indicates PAM, gray area/red dashed line indicates a deletion and pink indicates altered amino acids. (I) qRT-PCR for kif5Ba from pools of 5-dpf WT and homozygous mutants. kif5Ba undergoes nonsense-mediated decay in kif5Baae11/ae11 but not kif5Baae12/ae12 mutants. Error bars show mean±s.e.m.; Student′s t-test, *P=0.0388. |
|
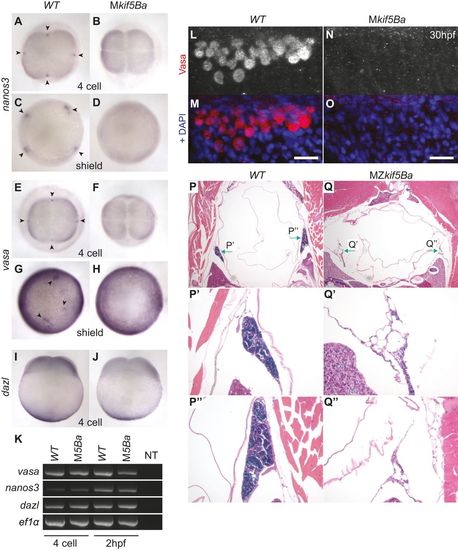
kif5Ba recruits germ plasm RNAs to cleavage furrows and promotes germ cell specification. (A,B,E,F) nanos3 (A,B) and vasa (E,F) localize to distal furrows in WT (A,E), but not in Mkif5Ba mutants (B,F), at 4-cell stage. Arrowheads indicate nanos3 and vasa accumulations. [nanos3: n=1/28 for Mkif5Baae11/ae12 (N=2 females), n=0/15 for Mkif5Baae11/ae11 (N=1 female), n=30/36 for WT (N=2 females) embryos have signal at ≥1 furrow; vasa: n=0/17 for Mkif5Baae11/ae12 (N=1 female), n=0/19 for Mkif5Baae11/ae11 (N=1 female), n=0/12 for Mkif5Baae12/ae12 (N=1 female), n=31/31 for WT (N=2 females) embryos have signal at e1 furrow.] (C,D,G,H) nanos3 (C,D) and vasa (G,H) are enriched in PGCs in WT (C,G), but not in Mkif5Ba mutants (D,H) at shield stage. Arrowheads indicate PGCs. [nanos3: n=0/32 for Mkif5Baae11/ae12 (N=2 females), n=0/19 for Mkif5Bae1/e1 (N=1 female), n=28/28 for WT (N=2 females) embryos have PGCs; vasa: n=0/18 for Mkif5Baae11/ae12 (N=1 female), n=0/22 for Mkif5Baae11/ae11 (N=1 female), n=0/17 for Mkif5Baae12/ae12 (N=1 female), n=37/37 for WT (N=2 females) embryos have PGCs.] (I,J) Vegetal localization of dazl is intact in WT (I) and Mkif5Ba mutants (J) at 4-cell stage. [n=19/19 for Mkif5Baae11/ae12 (N=2 females), n=20/20 for WT (N=2 females) embryos have vegetal signal.] (K) RT-PCR shows that germ plasm RNAs vasa, nanos3 and dazl are inherited by WT and Mkif5Ba mutant eggs (4-cell stage) and are maintained in WT and Mkif5Ba embryos (2hpf). ef1α, loading control. NT, no template. (L-O) Vasa+ PGCs are present in WT (L,M) but not in Mkif5Ba mutant (N,O) embryos at 30hpf. Scale bars: 10µm. [n=0/48 for Mkif5Baae11/ae12 (N=2 females), n=19/67 for Mkif5Baae11/ae11 (N=3 females), n=0/20 for Mkif5Baae12/ae12 (N=3 females), n=53/53 for WT (N=4 females) have PGCs.] (P-Q3) H&E-stained transverse sections of adult WT (P) and MZkif5Ba mutants (Q) show that MZkif5Ba testes (green arrows) lack germ cells. P′,Q′ and P′′,Q′′ are higher magnification images of left and right testes, respectively. [n=4/4 Mkif5Baae12/ae12 adult males had empty testes; in addition, n=18/18 adult Mkif5Baae11/ae12 males dissected had testes devoid of germ cells.] |
|
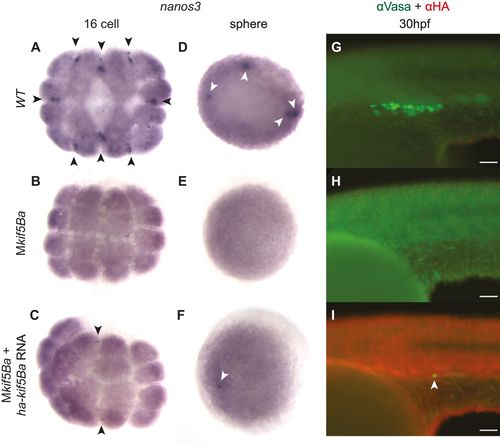
Overexpression of kif5Ba in Mkif5Ba mutants following fertilization is sufficient to specify germ cells. (A-C) nanos3 RNA is recruited to the first three cleavage furrows in WT (A) but is not recruited to furrows in Mkif5Ba mutants (B) at 16-cell stage. Injection of ha-kif5Ba restores nanos3 recruitment to 1-2 furrows in a fraction of Mkif5Ba mutants (C). Black arrowheads denote recruited nanos3. (D-F) nanos3 RNA becomes enriched in the prospective PGCs of WT (D) but is not enriched in Mkif5Ba mutants (E). Injection of ha-kif5Ba restores nanos3 enrichment to one PGC in a fraction of Mkif5Ba mutants (F). White arrowheads denote PGCs. (G-I) Vasa+ PGCs are present in WT (G) but not in Mkif5Ba mutants (H) at the yolk-yolk extension intersection. Injection of ha-kif5Ba restores small numbers of PGCs in Mkif5Ba mutants (arrowhead in I). See text for quantification. Scale bars: 50µm. |
|
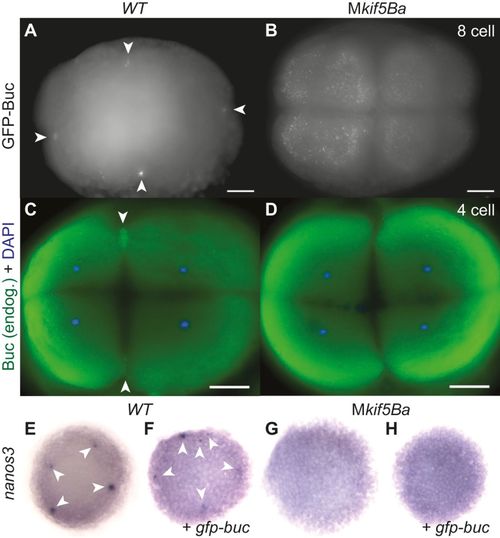
Kif5Ba localizes Buc to cleavage furrows to mediate germ plasm assembly in the embryo. (A,B) GFP-Buc is recruited to distal furrows of WT (arrowheads in A) but not of Mkif5Ba mutants (B). [n=0/29 for Mkif5Baae11/ae12 (N=2 females), n=0/18 for Mkif5Baae11/ae11 (N=3 females), n=0/15 for kif5Baae12/ae12 (N=1 female), n=32/32 for WT (N=4 females) have compact distal GFP-Buc; n=2/29 for Mkif5Baae11/ae12, n=15/18 for Mkif5Baae11/ae11, n=0/15 for Mkif5Baae12/ae12 have some furrow recruitment of GFP-Buc, although distal furrow compaction is defective.] (C,D) Endogenous Buc localizes to distal cleavage furrows of WT (arrowheads in C) but not Mkif5Ba mutants (D). [n=2/15 for Mkif5Baae12/ae12 (N=2 females), n=2/17 for Mkif5Baae11/ae11 (N=1 female), n=24/32 for WT (N=4 females) of cells with decondensed chromosomes have Buc at e1 distal cleavage furrow.] (E-H) Compared with uninjected WT (E), gfp-buc injected WT (F) embryos have additional nanos3+ PGCs (arrowheads), whereas uninjected (G) and gfp-buc-injected (H) Mkif5Ba mutants lacked nanos3+ cells. See text for quantification. |
|
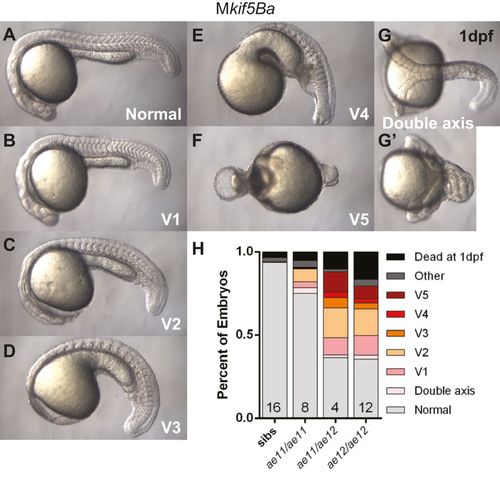
Maternal kif5Ba is required for dorsal fate. (A-G) Mkif5Ba mutant embryos are variably ventralized at 1dpf: V1, nearly complete AP axis lacking notochord and blocky somites (B); V2, reduced head structures (C); V3, largely lacking head structures except the posteriormost ones (D); V4, lack all head structures but possess an AP axis (E); V5, completely radialized axis (F). A small fraction of mutant embryos had duplicated anterior axes with fused caudal bodies (G,G2). (H) Distribution of phenotypic classes for homozygous and transheterozygous Mkif5Ba mutant embryos. The number of females assayed is indicated at the base of each bar. At least two clutches of embryos from each female were analyzed. Total number of embryos assayed for each genotype: n=1247 for Mkif5Baae11/ae12, n=2748 for Mkif5Baae11/ae11, n=2158 for Mkif5Baae12/ae12 and n=988 for siblings. PHENOTYPE:
|
|
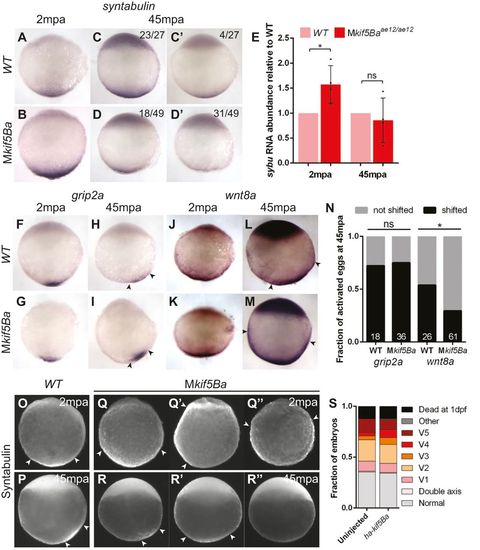
Maternal kif5Ba localizes dorsal determination components in activated eggs. (A-D2) sybu is vegetally localized at 2mpa (A,B) and at 45mpa (C,D) in both WT (A,C) and Mkif5Ba mutant (B,D) activated eggs. However, sybu is more dispersed at 2mpa (B) and appears less abundant in most Mkif5Ba eggs at 45mpa (D,D′). [2mpa: n=30/30 for Mkif5Baae11/ae12 (N=2 females), n=29/29 for Mkif5Baae12/ae12 (N=1 female), n=63/63 for WT (N=3 females) have vegetally localized sybu; 45mpa: quantification in panels.] (E) qRT-PCR reveals significantly more abundant sybu RNA in Mkif5Ba mutants at 2mpa, although this RNA excess is not maintained at 45mpa. Error bars show mean±s.d.; Student′s t-test, *P=0.0219. (F-I) grip2a is vegetally localized at 2mpa (F,G) and shifts asymmetrically by 45mpa (H,I) in both WT (F,H) and Mkif5Ba mutant (G,I) activated eggs. [2mpa: n=37/37 for Mkif5Baae11/ae12 (N=2 females), n=23/23 for WT (N=2 females) have vegetally localized grip2a; 45mpa: see N for quantification.] (J-M) wnt8a is vegetally localized at 2mpa in both WT (J) and Mkif5Ba mutant (K) activated eggs. However, wnt8a is asymmetric at 45mpa in WT (L) but not Mkif5Ba mutant (M) activated eggs. [2mpa: n=24/24 for Mkif5Baae11/ae12 (N=2 females), n=29/29 for WT (N=2 females) have vegetally localized wnt8a; 45mpa: see N for quantification.] (N) Fraction of eggs exhibiting dorsally translocated RNA at 45mpa. Number at base of columns indicates the number of eggs assayed. Chi-square, P=0.0312 (grip2a: n=2 females for each genotype; wnt8a: n=4 Mkif5Ba and n=3 WT females). (O-′′) Sybu is vegetally localized at 2mpa (O) and is asymmetric at 45mpa (P) in WT activated eggs. This is normal in some Mkif5Ba mutant eggs (Q,R), but others show aberrant localization at 2mpa (Q′,Q′′) and at 45mpa (R′,R′′). See text for quantification. (S) Injection of ha-kif5Ba RNA into 1-cell-stage Mkif5Ba mutants fails to suppress DV patterning defects. [n=138 uninjected and n=81 injected Mkif5Baae11/ae12 (N=1 female), n=172 uninjected and n=80 injected Mkif5Baae11/ae12 (N=2 females)]. Arrowheads designate the lateral limits of each expression domain. |
|
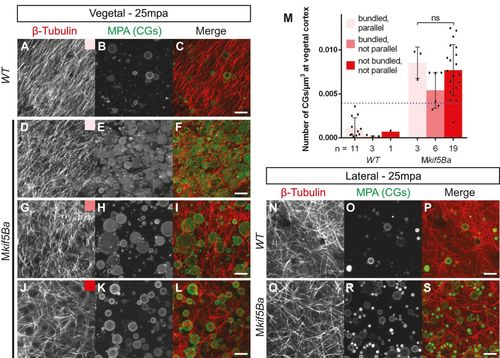
Maternal kif5Ba organizes vegetal microtubules. (A-M) Vegetal microtubules are in a parallel orientation and bundled in WT activated eggs (A), but in Mkif5Ba mutants are either bundled and parallel (D), bundled but not parallel (G), or not bundled or parallel (J). Mkif5Ba mutant activated eggs display variable CG exocytosis defects (compare B with E,H,K). Importantly, the degree of CG retention does not correlate with the severity of the microtubule phenotype in Mkif5Ba mutants (M). Scale bars: 10µm. Blue dotted line in M represents the upper limit of CG retention observed in WT activated eggs. Error bars show mean±s.d.; one-way ANOVA. (N-S) There is no difference between lateral microtubule organization in WT (N-P) and Mkif5Ba mutant (Q-S) activated eggs. Scale bars: 10µm. |
|
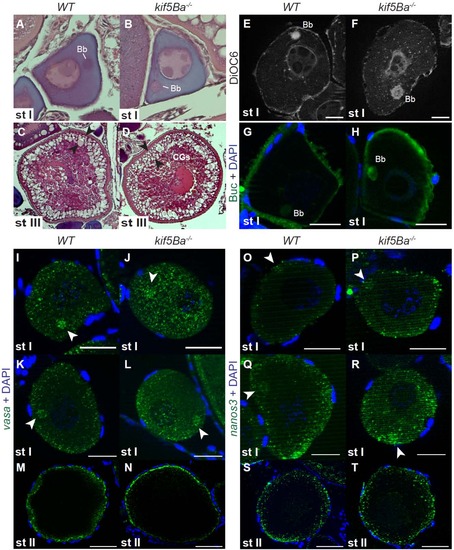
kif5Ba is dispensable for oocyte polarity and germ plasm localization. (A-D) H&E staining of WT (A,C) and kif5Ba-/- mutant (B,D) oocytes showing polarized stage I (st I) oocytes with Balbiani bodies (Bb) and normal stage III (st III) oocytes with cortically-localized cortical granules (CGs, between arrowheads) and centrally localized nucleus and yolk. (n=3 mutant ovaries analyzed, one each of kif5Baae11/ae11, kif5Baae11/ae12, and kif5Baae12/ae12; assayed 5-10 oocytes for each genotype). (E-F) DiOC6 staining reveals that ER and mitochondria are properly localized to the Bb in WT (E) and kif5Ba-/- mutant (F) st I oocytes. (n=18 oocytes analyzed from kif5Baae11/ae11 mutant ovary, n=19 oocytes analyzed from WT ovary). Scale bars 20µm. (G-H) Buc is properly localized to the Bb in WT (G) and kif5Ba-/- mutant (H) st I oocytes. (n=12 oocytes analyzed from kif5Baae11/ae11 mutant ovary, n=10 oocytes analyzed from WT ovary)). Scale bars 20µm. (I-T) vasa (I-N) and nanos3 (O-T) RNA are enriched in the Bb (arrowheads) in early st I (I,J,O,P) and late st I (K,L,Q,R) oocytes in WT (I,K,O,Q) and kif5Ba-/- mutants (J,L,P,R). vasa RNA localizes to the cortex in WT (M) and kif5Ba-/- mutant (N) st II oocytes. nanos3 RNA is diffuse throughout the cytosol of WT (S) and kif5Ba-/- mutant (T) st II oocytes. (n=3 WT and kif5Baae12/ae12 mutant ovaries, assayed ≥10 oocytes for each condition). Scale bars 25µm (I-L, O-R), 25µm (M,N,S,T). |
|
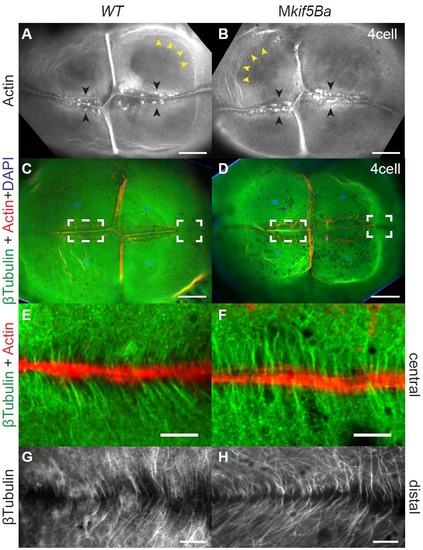
Cleavage furrow-associated cytoskeleton is intact in Mkif5Ba mutants. (A-B) Rhodamine-phalloidin staining to label F-actin. Animal pole views reveal an Factin contractile band present in all four furrows, F-actin accumulations flanking the FMA (black arrowheads) and circumferential actin bands around the blastomeres (yellow arrowheads) in both WT (A) and Mkif5Ba mutants (B). Scale bars, 100µm. (C-D) β-Tubulin antibody staining to reveal microtubules. Animal pole views show the Factin contractile band present in all four furrows and the furrow microtubule arrays that flank the newly forming furrows of WT (C) and Mkif5Ba mutants (D). Left and right dotted boxes in each panel indicate central and distal regions of the newly forming furrow respectively. Scale bars, 100µm. (E-F) Higher magnification of the central furrow shows that the furrow microtubules are parallel to each other and perpendicular to the F-actin contractile band in both WT (E) and Mkif5Ba mutants (F). Scale bars, 10µm. (G-H) Higher magnification of the distal furrow shows microtubules angled in a Vshaped configuration pointing towards the distal end in both WT (G) and Mkif5Ba mutants (H). Scale bars, 10µm. (All results are from n=11 embryos for Mkif5Baae11/ae12, n=9 for Mkif5Baae11/ae11, and n=15 for WT). |
|
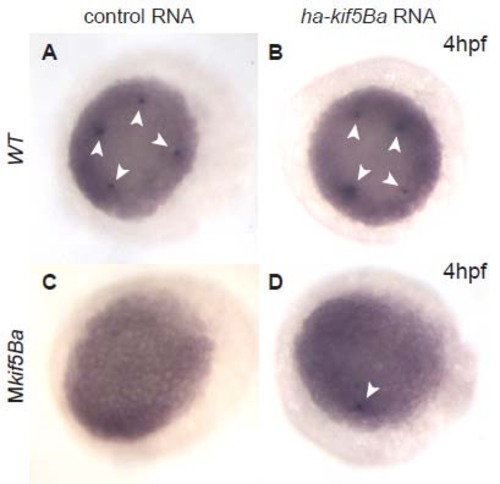
Injection of control RNA has no effect on number of PGCs in WT or Mkif5Ba mutant embryos. (A-B) The is no difference in the number of nanos3-positive PGCs at 4hpf in embryos injected with either 800pg of a control RNA encoding Cherry protein (A; 3.46±0.26 PGCs in n=37 embryos) or 800pg of ha-kif5Ba RNA (B; 3.56±0.25 PGCs in n=32 embryos) (C-D) While injection of 800pg of a control RNA encoding Cherry protein has no effect on the number of PGCs in Mkif5Ba mutants (C; zero PGCs in n=42/43 embryos, one PGC in n=1/43 embryos) injection of 800pg of ha-kif5Ba leads to the appearance of one PGC in a subset of Mkif5Ba mutants (D; one PGC visible in 13/85 embryos). Compare with uninjected Mkif5Ba mutants (zero PGCs in n=72/73 embryos, one PGC in n=1/73 embryos). |