- Title
-
Angiopoietin-like proteins stimulate HSPC development through interaction with Notch receptor signaling
- Authors
- Lin, M.I., Price, E.N., Boatman, S., Hagedorn, E.J., Trompouki, E., Satishchandran, S., Carspecken, C.W., Uong, A., DiBiase, A., Yang, S., Canver, M.C., Dahlberg, A., Lu, Z., Zhang, C.C., Orkin, S.H., Bernstein, I.D., Aster, J.C., White, R.M., Zon, L.I.
- Source
- Full text @ Elife
|
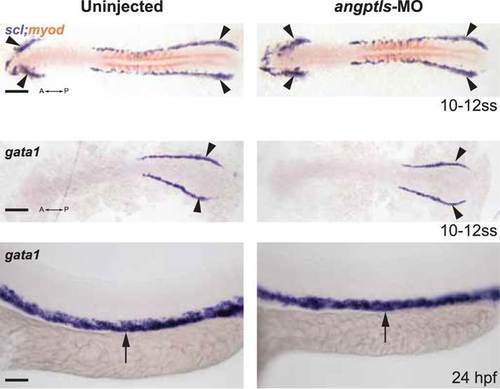
Angptls-MO morphants had no defect in primitive hematopoiesis. Top panels: somite matched embryos are scored by double staining with myod (orange, staining somite boundaries) and scl (purple, for early blood and vascular progenitor cells in the anterior (A) and posterior (P) bilateral stripes of the lateral plate mesoderm (LPM), black arrowheads, 10–12 ss). Middle and bottom panels: gata1-positive proerythroblast formation was also unaffected in these morphants in the posterior LPM (black arrowheads, 10–12 ss) and intermediate cell mass (black arrows, 24hpf). Scale bars: 200 µm (for top and middle panels); 50 µm (for bottom panels). |
|
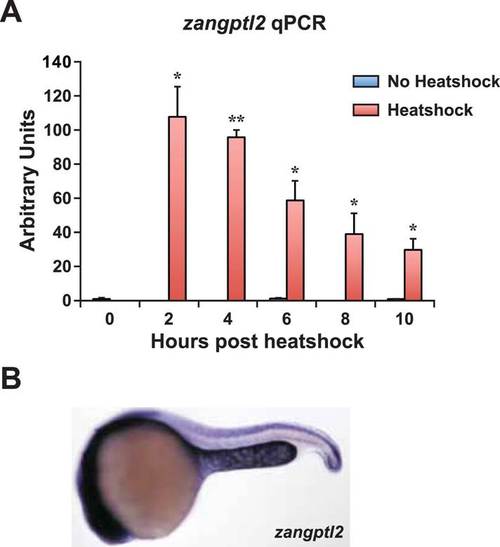
Zangptl2 overexpression in Tg(hsp70:zangptl2) embryos and endogenous zangptl2 expression. (A) qPCR analysis of zangptl2 mRNA levels in Tg(hsp70:zangptl2) embryos that have been heatshocked for 1 hr and collected at the indicated times post-heatshock. Heatshocked embryos (red bars) overexpressed zangptl2 mRNA at least 100-fold in excess compared to non-heatshocked siblings (blue bars). Error bars denote S.E.M., *p < 0.05, **p < 0.01 compared to 0 hr, one way ANOVA. (B) WISH of endogenous zangptl2 at 23hpf (the highest of all timepoints observed) is mostly restricted in the yolk sac extension, spinal cord, and head region. |
|
Angptls-MO morphants had no defect in primitive hematopoiesis. Top panels: somite matched embryos are scored by double staining with myod (orange, staining somite boundaries) and scl (purple, for early blood and vascular progenitor cells in the anterior (A) and posterior (P) bilateral stripes of the lateral plate mesoderm (LPM), black arrowheads, 10–12 ss). Middle and bottom panels: gata1-positive proerythroblast formation was also unaffected in these morphants in the posterior LPM (black arrowheads, 10–12 ss) and intermediate cell mass (black arrows, 24hpf). Scale bars: 200 µm (for top and middle panels); 50 µm (for bottom panels). |
|
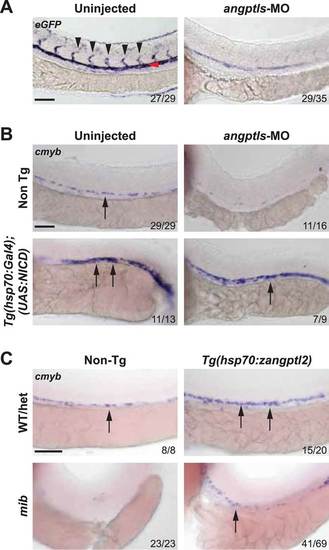
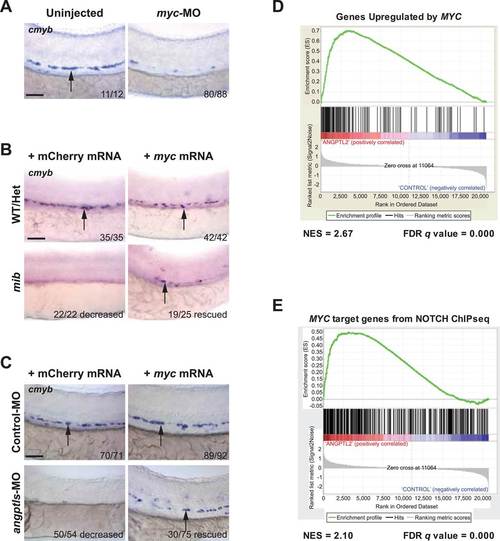
Angptls genetically interact with notch. (A) Notch eGFP Tg reporter embryos injected with angptls-MO (2 ng) had decreased notch activity in ISV (black arrowheads) and DA (red arrowhead) at 28hpf. (B) Overexpression of constitutively active notch in angptls-MO injected (2 ng) Tg(hsp70:Gal4;UAS:NICD) can restore cmyb-positive HSPCs (arrows) at 36hpf after heatshock. (C) Mib had no cmyb-positive HSPCs (arrows) at 36hpf compared to WT or het siblings. Overexpression of zangptl2 in mib by crossing Tg(hsp70:zangptl2) into mib can restore this defect upon heatshock. Scale bars: 50 µm. |
|
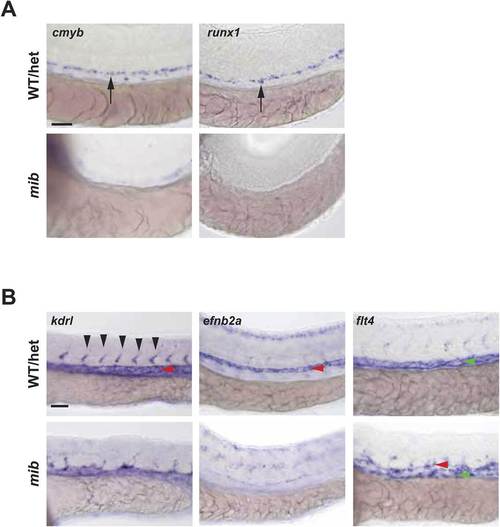
Notch mutant mib has hematopoietic and vascular defects. (A) Mib has no cmyb- or runx1-positive HSPCs (arrows) in the AGM at 36hpf. (B) Mib embryos also display vascular defects including disorganized ISV sprouting (black arrowheads), loss of arterial marker, efnb2a in the DA (red arrowheads), and ectopic expression of the venous marker, flt4 in the DA in addition to the PCV (green arrowheads) at 28hpf. Scale bars: 50 µm. |
|
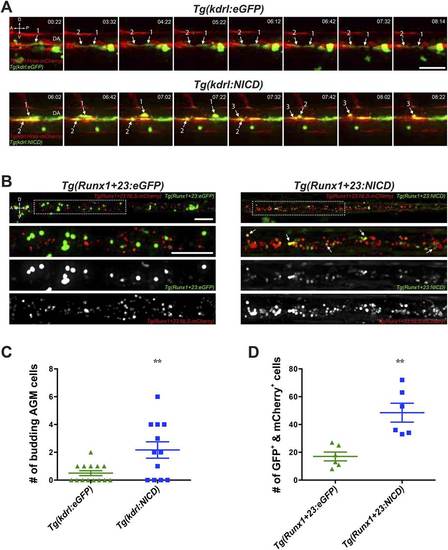
Notch cell autonomously increase definitive hematopoiesis. (A) Time-lapse sequence (hours:minutes post-28hpf) of HSPCs emerging from the ventral wall of the DA. Hemogenic endothelial cells (red) that have incorporated the injected transgene (green) are marked with numbers and white arrows. Each injected embryo was scored for 24 hr and tabulated in (C). Scale bar: 25 µm. (B) Still images of the CHT from 72hpf embryos. Runx1-positive HSPCs (red) that have incorporated the injected transgene (green) were scored for double positivity (yellow, examples marked by white arrows) and tabulated in (D). Boxed areas in the top panels are magnified and split into double or single fluorescent panels below. Scale bars: 50 µm. A: Anterior, P: Posterior, D: Dorsal; V: Ventral. Error bars denote S.E.M., **p < 0.01, compared to eGFP injected controls, Student′s t test. |
|
NOTCH ChIP-seq in ANGPTL2-stimulated CD34+ cells. (A) Enriched transcription factor motifs at the peaks of NOTCH binding sites from NOTCH ChIP-seq. Parentheses indicate E values for the significance of the motif. An embedded RBPjK ((consensus motif (red box) was found within the ZNF143 motif as previously described (Wang et al., 2011). (B) Examples of MYC-related categories from GREAT analysis of NOTCH ChIP-seq. (C) IPA network analysis of NOTCH ChIP-seq bound genes revealed a dense network of target genes centered on MYC. |