- Title
-
Leptin and IL-6 Family Cytokines Synergize to Stimulate Müller Glia Reprogramming and Retina Regeneration
- Authors
- Zhao, X.F., Wan, J., Powell, C., Ramachandran, R., Myers, M.G., Goldman, D.
- Source
- Full text @ Cell Rep.
|
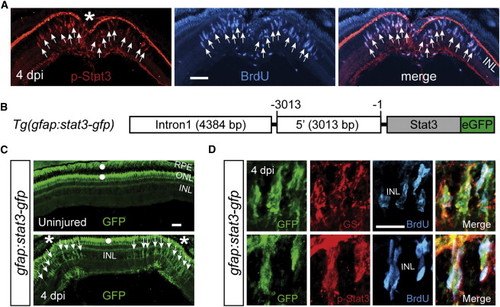
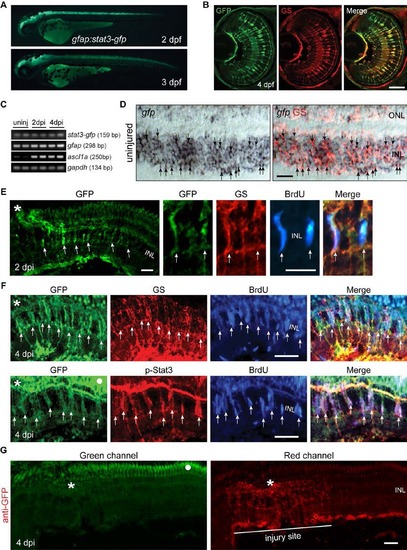
The Jak/Stat3 Signaling Pathway Is Activated following Retinal Injury (A) Immunofluorescence on retinal sections shows activated p-Stat3 expression in BrdU+ MG-derived progenitors that are localized to the injury site at 4 dpi. (B) A schematic of the gfap:stat3-gfp transgene construct shows the fusion gene, stat3-gfp, under control of the gfap promoter regulatory elements. (C) In gfap:stat3-gfp transgenic fish, Stat3-GFP fusion protein expression is undetectable in MG of the uninjured eye and is restricted to MG-derived progenitors at the injury site at 4 dpi. White dots indicate autofluorescence unique to the green channel (see Figure S2G). (D) Confocal images show colocalization of Stat3-GFP with GS+/p-Stat3+/BrdU+ MG-derived progenitors at 4 dpi. In (A) and (C), the asterisk marks the injury site (needle poke) and arrows point to MG-derived progenitors. Scale bars, 50 µm (A and C) and 20 µm (D). INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium; dpi, days postinjury. See also Figures S1 and S2. EXPRESSION / LABELING:
|
|
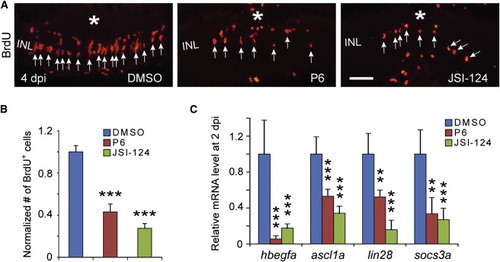
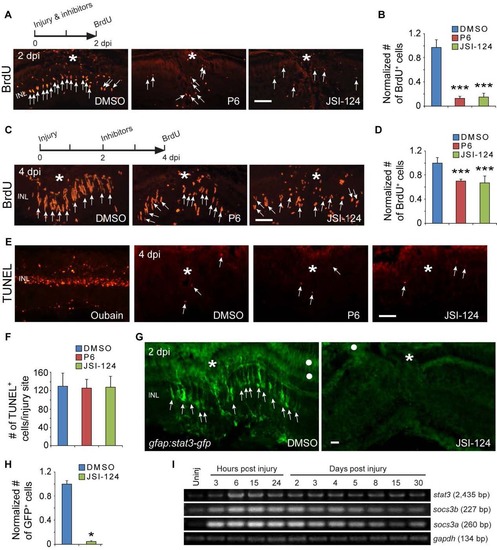
The Jak/Stat3 Signaling Pathway Is Necessary for Retina Regeneration (A) BrdU immunofluorescence shows that the Jak inhibitors P6 and JSI-124 suppress progenitor formation at 4 dpi. The asterisk marks the injury site (needle poke) and arrows point to MG-derived progenitors. (B) Quantification of BrdU+ progenitors in (A). p < 0.001, n = 4. (C) qPCR shows that the Jak inhibitors P6 and JSI-124 inhibit reprogramming gene induction at 2 dpi; p < 0.01, p < 0.001; n = 4, 4, and 5 for DMSO, P6, and JSI-124, respectively. Error bars, SD. Scale bars, 20 µm (A). See also Figure S3. PHENOTYPE:
|
|
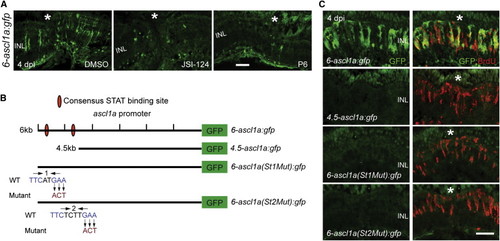
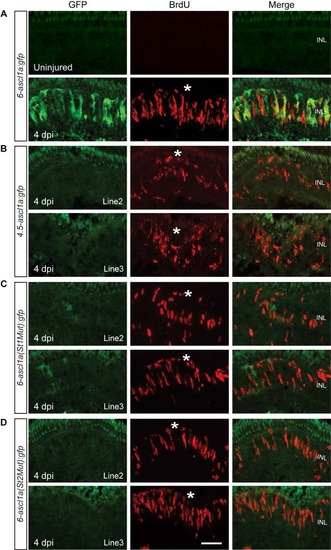
Jak/Stat3 Signaling Mediates Injury-Dependent Induction of the Reprogramming Gene ascl1a (A) GFP immunofluorescence in 6-ascl1a:gfp fish shows that the Jak inhibitors P6 and JSI-124, applied at the time of retinal injury, inhibit injury-dependent transgene induction. (B) Diagram of the ascl1a promoter constructs used to generate transgenic lines. (C) GFP immunofluorescence shows that a distal 1.5 kb fragment of the ascl1a promoter is required for injury-dependent transgene expression and that both consensus Stat3 sites located in this promoter fragment are necessary for this expression. BrdU+ cells indicate the injury site and a normal regenerative response. The asterisk marks the injury site (needle poke). Scale bar, 50 µm. See also Figure S4. EXPRESSION / LABELING:
|
|
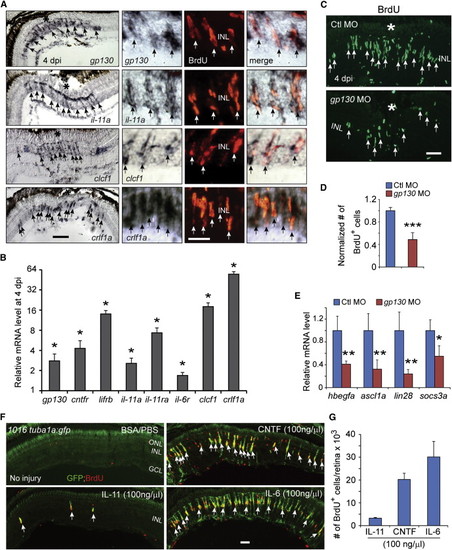
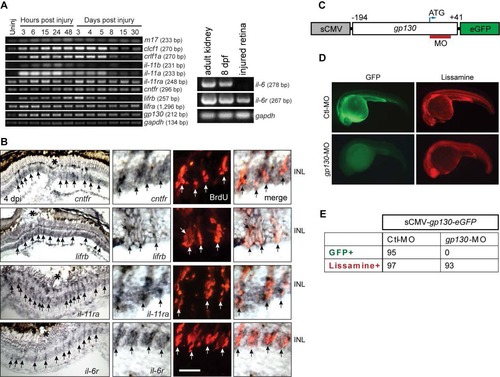
IL-6 Family Cytokines Signaling through Gp130 Are Necessary and Sufficient for Retina Regeneration (A) In situ hybridization and immunofluorescence show that gp130, il-11a, crlf1a, and clcf1 are expressed in BrdU+ MG-derived progenitors localized to the injury site. (B) qPCR quantifies il-6 family gene induction in MG-derived progenitors (FACS purified from 1016tuba1a:gfp fish retinas at 4 dpi) relative to MG from uninjured retina (FACS purified from uninjured gfap:gfp fish retinas). p < 0.05, n = 3. (C and D) Gp130 knockdown inhibits the generation of BrdU+ MG-derived progenitors at 4 dpi. Control (Ctl) or gp130-targeting MOs were electroporated into the retina at the time of injury and the fish received an i.p. injection of BrdU 3 hr before sacrifice on 4 dpi. p < 0.001, n = 4. (E) qPCR showing Gp130 knockdown inhibits injury-dependent induction of reprogramming genes at 2 dpi. p < 0.05, p < 0.01, n = 4. (F) Intravitreal injection of recombinant mammalian IL-6-like cytokines into the uninjured eye of 1016tuba1a:gfp fish stimulates GFP expression and BrdU incorporation in MG throughout the retina’s INL. Intravitreal injection of PBS/BSA did not stimulate GFP expression or BrdU incorporation. The green fluorescence above the ONL in the top left-hand panel is autofluorescence unique to the green channel (see Figure S7K). (G) Quantification of BrdU+ cells following intravitreal injection of recombinant mammalian IL-6-like cytokines (n = 3). Error bars, SD. In (A) and (C), the asterisks mark the injury site (needle poke). In (A), (C), and (F), arrows point to MG-derived progenitors. Scale bars, 20 µm (A and C) and 50 µm (F). GCL, ganglion cell layer. Primers are listed in Table S1. See also Figures S5 and S6. |
|
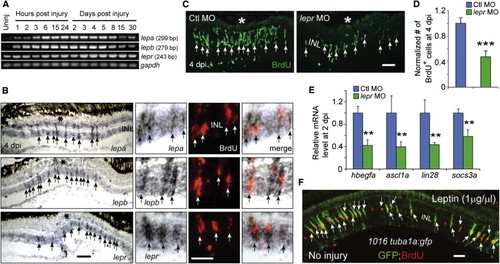
Leptin Signaling Is Necessary and Sufficient for Retina Regeneration (A) RT-PCR analysis of mRNAs (from whole retina) encoding Leptin and Lepr at various times after retinal injury. (B) In situ hybridization and BrdU immunofluorescence shows that lepa, lepb, and lepr RNAs are increased in BrdU+ MG-derived progenitors at the injury site. The asterisk marks the injury site (needle poke). (C and D) MO-mediated knockdown of Lepr inhibits the generation of BrdU+ MG-derived progenitors at 4 dpi. p < 0.001, n = 4. (E) qPCR shows that Lepr knockdown suppresses injury-dependent induction of reprogramming genes at 2 dpi. p < 0.01, n = 4. (F) Intravitreal injection of recombinant human Leptin into the uninjured eye of 1016tuba1a:gfp fish stimulates GFP expression and BrdU incorporation in MG throughout the retina’s INL. Error bars, SD. In (B) and (C), arrows point to MG-derived progenitors. Scale bars, 20 µm (B and C) and 50 µm (E). Primers are listed in Table S1. See also Figure S7. |
|
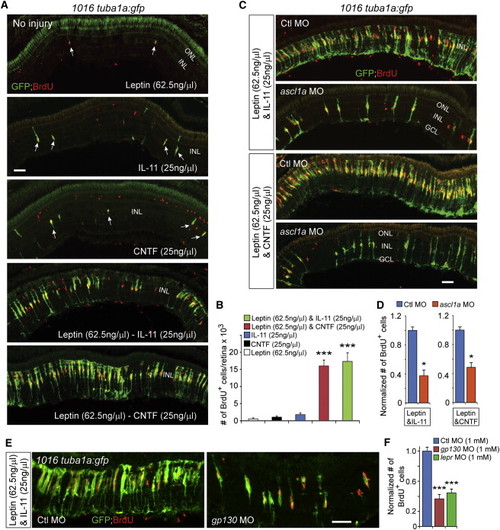
Leptin Synergizes with IL-11 and CNTF to Stimulate MG Reprogramming and Proliferation in the Uninjured Retina (A) GFP and BrdU immunofluorescence shows that Leptin synergized with IL-11 and CNTF to stimulate GFP expression and MG proliferation in the uninjured retina 1016tuba1a:gfp fish, whereas Leptin, IL-11, or CNTF alone had little effect. Arrows point to MG-derived progenitors in the top three panels. (B) Quantification of the effects of cytokines on MG proliferation when delivered individually or in combination to the uninjured retina. p < 0.001 (combination versus individual), n = 4 per group. (C and D) MO-mediated Ascl1a knockdown inhibits the synergistic effects of Leptin/IL-11 or Leptin/CNTF on GFP induction and MG proliferation. p < 0.05, n = 3. (E and F) Knockdown of Gp130 or Lepr inhibits the synergistic effects of Leptin/IL-11 on proliferation. p < 0.001, n = 4. Error bars, SD. Scale bars, 50 µm (A, C, and E). |
|
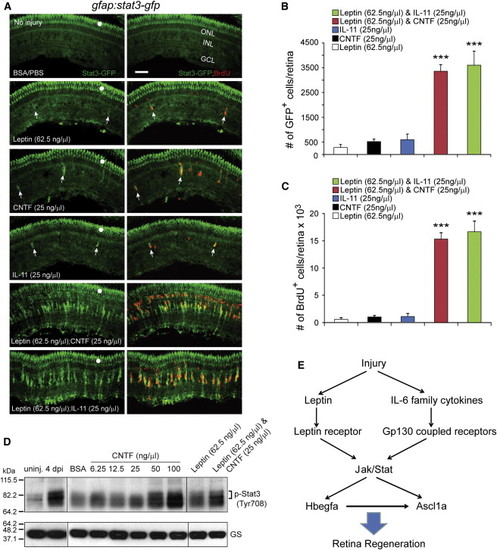
Leptin Synergizes with IL-11 and CNTF to Stimulate Jak/Stat3 Signaling in MG-Derived Progenitors (A) Intravitreal injection of Leptin/CNTF, or Leptin/IL-11 into the eye of gfap:stat3-gfp fish stimulated Stat3-GFP expression and BrdU incorporation throughout the uninjured retina’s INL, whereas Leptin, CNTF, or IL-11 alone had little effect. Note that GFP reports activated p-Stat3 expression (Figures 1 and S2). White dots indicate autofluorescence unique to the green channel (see Figure S7K). The arrows point to MG-derived progenitors in the top four panels. (B) Quantification of Stat3-GFP+ cells in (A). p < 0.001 (combination versus individual), n = 4. (C) Quantification of BrdU+ cells in (A). p < 0.001 (combination versus individual), n = 4. (D) Western blot shows that retinal injury or intravitreal injection of cytokines into an uninjured eye increases p-Stat3 expression. GS serves as the loading control. (E) Model showing that Leptin and IL-6 family cytokines synergize to stimulate MG reprogramming and retina regeneration via a Jak/Stat3 signaling pathway, which is essential for activating reprogramming genes such as hbegf and ascl1a. Error bars, SD. Scale bars, 50 µm. |
|
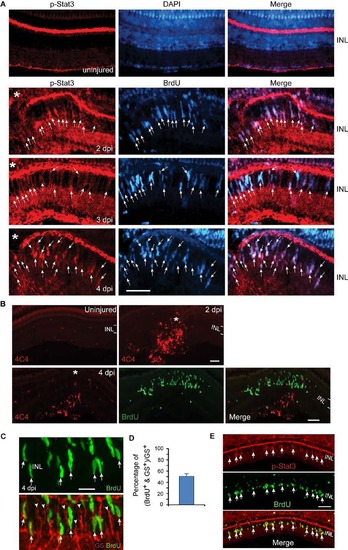
Injury-dependent activation of the Jak/Stat3 signaling pathway. Related to Figure 1. (A) p-Stat3 (red) and BrdU (blue) immunofluorescence shows p-Stat3 is induced in BrdU+ MG-derived progenitors at the injury site at 2-4 dpi; n=3. Arrows point to p-Stat3+/BrdU+ double labelled cells. (B) 4C4 immunoflourescence (red) shows microglia, diffusely scattered throughout the uninjured retina, accumulate at the injury site, but do not proliferate (lack BrdU co-labeling, green). The BrdU+ cells (green) are MG-derived progenitors confined to the INL. (C, D) Immunofluorescence shows ~50% of glutamine synthetase (GS)+ MG (red) incorporate BrdU+ 4 days after a 30 min exposure to UV light; n=3. Arrows point to BrdU+/GS+ double labelled cells. Arrowheads point to BrdU-/GS+ quiescent MG. (E) Immunofluorescence shows p-Stat3 is restricted to BrdU+ MG-derived progenitors 4 days after a 30 min exposure to UV light. Asterisks in (A) point to the injury site (needle poke). Scale bar, 20 µm (C); 50 µm (A,E). INL, inner nuclear layer. |
|
Stat3-GFP expression in developing and adult gfap:stat3-gfp fish. Related to Figure 1. (A) Stat3-GFP is expressed throughout brain and spinal cord in live gfap:stat3-gfp larva at 2 and 3 dpf. (B) Immunofluorescence on retinal sections shows co-localization of Stat3-GFP with glutamine synthetase (GS)+ MG at 4 dpf. (C) Whole retina RT-PCR showing constitutive gfp mRNA expression in the uninjured and injured adult retina, while ascl1a mRNA was induced after retinal injury. (D) In situ hybridization and immunofluorescence shows expression of gfp RNA in GS+ MG in adult uninjured retina (arrows). (E) Immunofluorescence shows Stat3-GFP expression co-localizes with GS +/BrdU+ MG-derived progenitors localized to the injury site at 2 dpi in the adult retina. (F) Immunofluorescence shows co-localization of Stat3-GFP with GS+/p-Stat3+/BrdU+ MG-derived progenitors at 4 dpi in the adult retina. White dot indicates autofluorescence. Asterisks mark the injury site (needle poke) and arrows point to MG-derived progenitors. (G) Anti-GFP immunofluorescence on a retinal section prepared at 4 dpi from gfap:stat3-gfp fish using a secondary antibody coupled to a red fluor shows autofluorescence in the ONL when viewed in the green channel (left-hand panel, dot marks autofluorescence) that is not evident in the red channel (right-hand panel). Asterisk marks the injury site. Scale bar, 50 µm (B, D-G). INL, inner nuclear layer; dpf, days post fertilization. |
|
Jak/Stat signaling regulates proliferation of MG-derived progenitors and Stat3-GFP expression. Related to Figure 2. (A, B) BrdU immunofluorescence on retinal sections shows that exposing fish to Jak inhibitors P6 or JSI-124 from 0-2 dpi inhibits progenitor proliferation at 2 dpi; ***P<0.001, n=4. (C, D) BrdU immunofluorescence shows that exposing fish to Jak inhibitors P6 or JSI-124 from 2-4 dpi inhibits progenitor proliferation at 4 dpi; ***P<0.001, n=4. (E, F) TUNEL staining for apoptotic cells at 4 dpi in retinas treated with DMSO, P6 or JSI-124. Oubain was used as a positive control. In panel F, n=3. (G, H) GFP immunofluorescence shows that exposing gfap:stat3-gfp fish to Jak inhibitors JSI-124 from 0-2 dpi inhibits Stat3-GFP expression; *P<0.05, n=3. (I) RT-PCR analysis of retinal stat3 and socs3 gene expression at various times after injury. Asterisks mark the injury sites (needle poke) and arrows point to MG-derived progenitors. White dot marks regions with autofluorescence. Error bars, s.d. Scale bar, 20 µm (A, C, E, G). INL, inner nuclear layer. |
|
Jak/Stat3 signaling mediates injury-dependent activation of the ascl1a promoter. Related to Figure 3. (A) GFP and BrdU immunofluorescence co-localize beneath the injury site in 6-ascl1a:gfp fish at 4 dpi. (B) A distal 1.5 kb ascl1a promoter fragment is necessary for injury-dependent transgene induction. (C, D) Consensus Stat3 sites located within this distal 1.5 kb region are necessary for injury-dependent transgene expression. BrdU+ immunofluorescence identifies the injury site and a normal regenerative response. The asterisks mark the injury sites. See Figure 3 for promoter schematic. Scale bar, 50 µm. INL, inner nuclear layer. |
|
Genes encoding IL-6 family cytokines are expressed in MG-derived progenitors upon retinal injury. Related to Figure 4. (A) RT-PCR analysis of retinal gene expression at various times after injury. il-6 is expressed in adult kidney and 8 dpf larva, but undetectable in the injured retina. (B) In situ hybridization shows induction of il-6 family member genes in BrdU+ MG-derived progenitors at the injury site. The asterisks mark the injury sites and the arrows point to MG-derived progenitors. Scale bars, 20 µm. (C) Diagram of sCMV:gp130-egfp reporter in which a fragment of gp130 DNA, containing the MO target sequence, is in-frame with the egfp coding sequence and under control of the sCMV promoter. (D) Injection of the sCMV:gp130-egfp reporter together with either lissamine-tagged control (Ctl) MO or gp130-targeting MO into one cell stage of zebrafish embryos and examined by fluorescence 24 hours later. Red fluorescence shows embryos received lissamine-tagged MOs. (E) Quantification of GFP-expressing embryos at 24 hpf. No GFP was detected in the gp130-targeting MO injected group, while GFP was readily detected in the control group. Similar results were obtained in 3 independent experiments. dpf, days post fertilization ; INL, inner nuclear layer. |
|
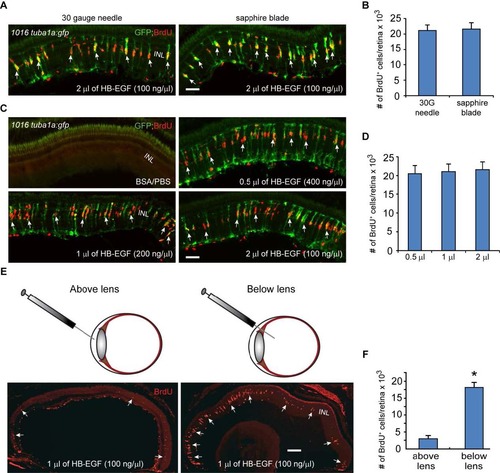
HB-EGF stimulates MG reprogramming and proliferation in the uninjured retina. Related to Figure 4. (A) A corneal puncture with a 30 gauge needle or a corneal incision made with a sapphire blade was followed by intravitreal injection of recombinant HB-EGF into the uninjured eye of 1016tuba1a:gfp fish. HB-EGF stimulated GFP expression and BrdU incorporation in MG throughout the retina’s inner nuclear layer. No GFP expression or BrdU incorporation is observed in PBS/BSA-injected eyes. (B) Quantification of BrdU+ cells in (A) shows similar effect of intravitreally delivered HB-EGF on MG proliferation when the cornea was either punctured with a needle or cut with a sapphire blade; n=3 per group. (C) A sapphire blade was used to make a small incision in the cornea and 200 ng of HB-EGF in 0.5 to 2 µl volumes was intravitreally injected into uninjured eyes of 1016tuba1a:gfp fish. (D) Quantification of BrdU+ cells following intravitreal delivery of different HB-EGF volumes reveals a similar effect on MG proliferation regardless of injection volume; n=3 per group. (E, F) HB-EGF must be delivered below the lens to stimulate MG proliferation; *P<0.05, n=3. Arrows point to MG-derived progenitor. Error bars, s.d. Scale bar, 50 µm (A, C); 150 µm (E). INL, inner nuclear layer. |
|
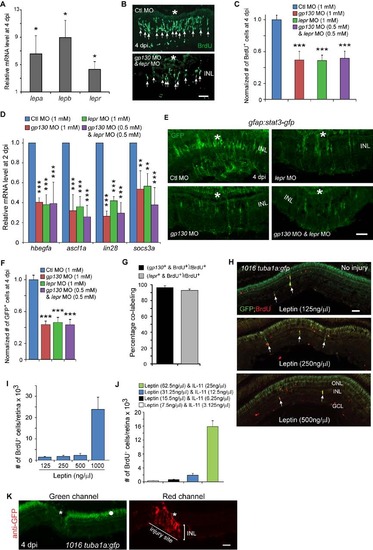
Leptin signaling regulates the generation of MG-derived progenitors. Related to Figure 5. (A) qPCR quantification of injury-dependent gene induction in FACS purified GFP+ MG-derived progenitors from the retina of 1016tuba1a:gfp fish at 4 dpi in comparison to that in FACS purified GFP+ MG from the uninjured retina of gfap:gfp fish; *P<0.05, n=3. (B) BrdU immunofluorescence shows that Leptin receptor and Gp130 knockdown inhibits the generation of BrdU+ MG-derived progenitors at 4 dpi. Asterisks mark the injury sites and the arrows point to MG-derived progenitors. (C) Quantification of the effects Gp130 and Leptin receptor knockdown have on progenitor formation (Figure 4C; Figure 5C); ***P<0.001, n=4. (D) qPCR showing Gp130 and Leptin receptor knockdown, individually or in combination, inhibits injury-dependent induction of reprogramming genes at 2 dpi; **P<0.01, ***P<0.001, n=4. (E) Knockdown of Gp130 inhibits the expression of Stat3-GFP at 4 dpi. (F) Quantification of the number of GFP+ cells in the retina at 4 dpi following Gp130 or Leptin knockdown; ***P<0.001, n=4. Error bars, s.d. (G) Quantification of the number of BrdU+ cells expressing gp130 or lepr RNA in the retina at 4 dpi; n=3. Error bars, s.d. (H) Effects of different Leptin concentrations on the generation of BrdU+ MG-derived progenitors and transgene expression in the uninjured retina of 1016tuba1a:gfp fish. The arrows point to MG-derived progenitors. (I, J) Quantification of the effect different Leptin (I) or Leptin/IL-11 (J) concentrations have on progenitor formation in the uninjured retina; n=3. Error bars, s.d. (K) GFP immunofluorescence using a secondary antibody coupled to a red fluor shows autofluorescence in the green channel (left-hand panel, dot) that is localized to the ONL and pigment epithelium, while the red channel shows GFP expression restricted to the injury site (right-hand panel, asterisk). Scale bars, 20 µm (B); 50 µm (E, H, K). ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. |