- Title
-
Nom1 Mediates Pancreas Development by Regulating Ribosome Biogenesis in Zebrafish
- Authors
- Qin, W., Chen, Z., Zhang, Y., Yan, R., Yan, G., Li, S., Zhong, H., Lin, S.
- Source
- Full text @ PLoS One
|
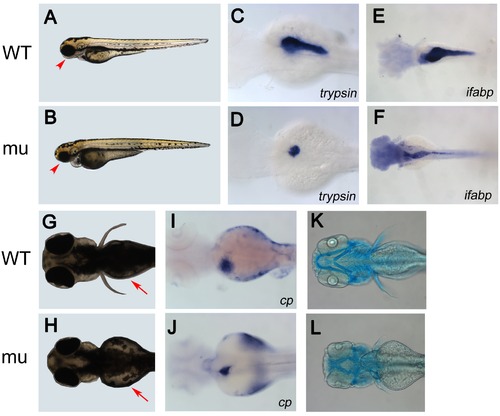
dg5 mutant has an endodermal and craniofacial defect. (A, B) Lateral and (G, H) dorsal views of live Wild Type (WT) sibling and dg5 larvae at 3 dpf. Smaller head and eyes can be seen (arrowhead in A, B). Impaired yolk absorption is apparent in dg5 mutant at 3 dpf (arrows in G, H). (C–F, I–L) All dorsal views, anterior to the left. At 3 dpf, the expression of trypsin (try) is markedly reduced in dg5 mutant (D) as compared to WT group (C). The intestine marker fatty acid binding protein 2 (ifabp) expression reveals that the intestine is thinner in dg5 larvae (E) as compared to WT group (F) at 3.5 dpf. The expression of ceruloplasmin (cp) shows that the liver is smaller in dg5 mutant (H) as compared to WT group (I) at 3 dpf. As compared to control group (K), craniofacial development is abnormal in dg5 larvae (L) by alcian blue staining at 4 dpf. EXPRESSION / LABELING:
PHENOTYPE:
|
|
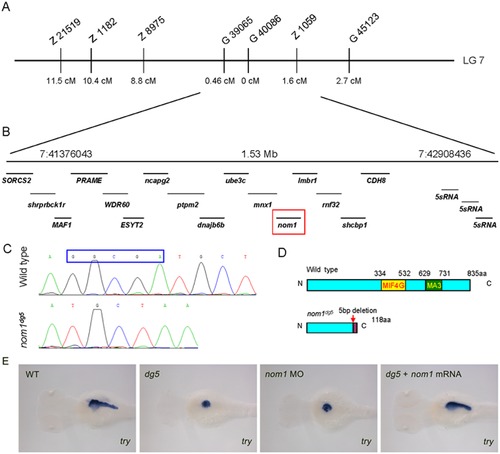
dg5 encodes zebrafish nom1. (A) Chromosome 7 is linked to the dg5 locus, the cM distance of different SSLP markers to dg5 locus is marked. (B) The region between markers G39065 and Z1059, contains 16 annotated genes (B). (C) Sequencing shows that nom1 from dg5 mutant has a 5-bp nucleotide deletion, producing a truncated nom1 protein. (E) nom1 ATG-MO injected embryos show the same decreased try expression level as compared to dg5 mutant and zebrafish nom1 mRNA can rescue try expression. Dorsal views, anterior to the left. EXPRESSION / LABELING:
PHENOTYPE:
|
|
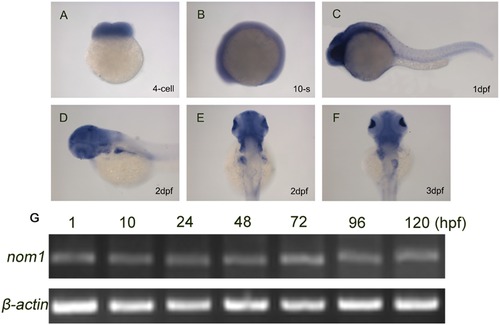
Nom1 expression in the developing zebrafish. (A–D) Lateral views, anterior to the left. (E, F) Dorsal views, anterior to the top. (A) nom1 has a strong maternal expression at 4-cell stage. (B–C) Non-specific ubiquitous nom1 expression is evident at 10-somite stage and 1 dpf. (D, E) At 2 dpf, nom1 expression in brain and liver is more apparent. (F) In addition to brain, nom1 is strongly expressed in pancreas and weakly in intestine at 3 dpf. (G) RT-PCR analysis shows that nom1 transcript is present at all embryonic stages. |
|
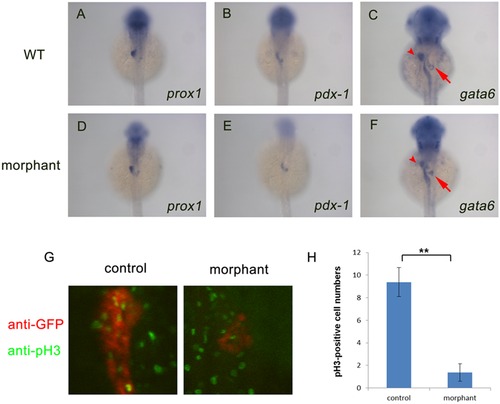
Nom1 is indispensable for pancreas proliferation but not for specification. (A–F) Dorsal views, anterior to the top. (A, B) Expression of prox1 (D, E) and pdx-1 (C, F) in the nom1 morphants is comparable to that in WT embryos at 36 hpf but reduced gata6 expression in liver (arrow) and pancreas (arrowhead) of nom1 morphants is obvious. (G) Anti-phospho Histone H3 (pH 3) staining for control and nom1 morphant embryos at 3 dpf. Red: GFP staining. Green: pH 3 staining. The signal of pH 3 staining in the pancreas of morphants is recognizably decreased. (H) Quantification of pH 3-positive cell numbers. Data were collected from 10 embryos. Error bars mean±SD. **P<0.01. EXPRESSION / LABELING:
PHENOTYPE:
|
|
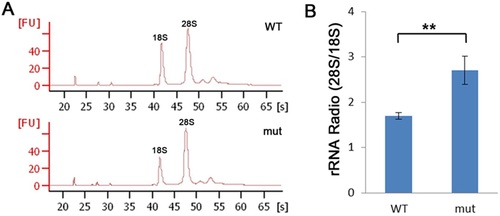
dg5nom1 larvae display defects in ribosome biogenesis. (A) Bioanalyser analysis of total RNA isolated from WT and mutant group at 3 dpf reveals a reduction of the 18S rRNA production but relative normal 28S rRNA amount. (B) The relative rRNA ratio (28S/18S) is elevated in the dg5nom1 as compared to WT. Error bars mean ± SD. **P<0.01. PHENOTYPE:
|
|
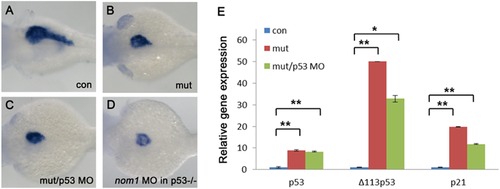
Nom1-induced pancreas defect is p53 independent. (A–D) Lateral views, anterior to the left. (A) control group at 3 dpf. (B) dg5 mutant group at 3 dpf. (C) Failure to rescue the pancreas defect by injection of 4 ng p53MO into dg5 mutant or (D) injection of nom1 MO in p53M214K mutant. (E) Expression of p53 and its targets Δ1113p53 and p21 are increased in dg5nom1 mutant, as assessed by quantitative PCR. Error bars mean±SD. **P<0.01, *P<0.05. EXPRESSION / LABELING:
PHENOTYPE:
|
|
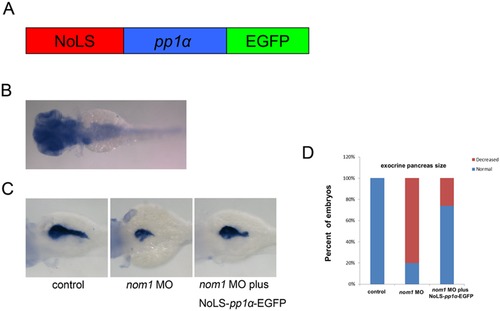
NoLS-pp1α-EGFP mRNA partially rescues the pancreas defect in nom1 morphant. (A) Schematic diagram of NoLS-pp1α-EGFP. Red: human nom1 NoLS. Blue: pp1α coding sequence. Green: EGFP coding sequence. (B) In situ result demonstrates that pp1α expresses in brain, liver, pancreas and intestine at 3 dpf. (C) Co-injection of NoLS-pp1α-EGFP mRNA and nom1 MO partially restores try expression as compared to nom1 morphant. (D) The percentage of embryos with relatively normal exocrine pancreas size is statistically higher in nom1 MO and NoLS-pp1α-EGFP co-injection group. |
|
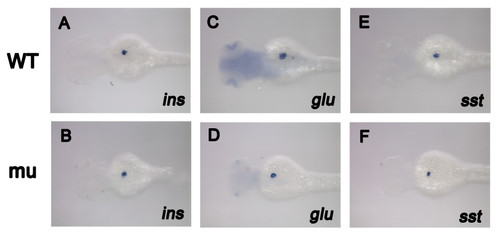
Endocrine pancreas markers expression was not affected in dg5 mutant larvae. (A, B) the endocrine pancreas β-cell marker ins was not affected in dg5 mutant at 3 dpf. (C–F) The same result can be seen in pancreas α-cell markern glu and pancreas δ–cell marker sst. All dorsal views, anterior to the left. |
|
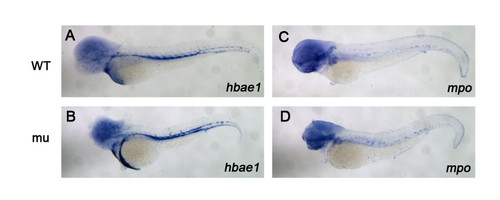
Dg5 larvae does not have an effect on hematopoiesis. (A–D) dg5 has a normal expression pattern on hemoglobin marker hbae1 and myeloid marker mpo compared to WT group. All lateral views, anterior to the left. |
|
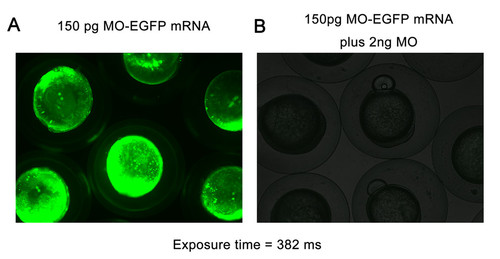
The efficiency verification of nom1 ATG MO. (A) Strong GFP fluorescence appeared in embryos injected with a fusion EGFP protein contained nom1 ATG MO target site. (B) The GFP fluorescence is disappeared in embryos co-injected with the fusion protein mRNA and 2 ng nom1 ATG MO. |
|
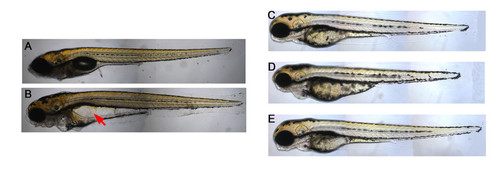
Phenotype of dg5 larvae at 7 dpf and the morphology of nom1-knockdown morphant and rescue embryos. (A, B) dg5 larvae do not have a swim bladder and cause a serious edema (arrow) at 7 dpf. (C) Compared to control embryos, (D) nom1 ATG-MO can cause the same phenotype as dg5 mutant with small head, small eyes and heart edema. (E) Injection nom1 mRNA into dg5 mutant can rescue the morphology phenotype. All lateral views, anterior to the left. |
|
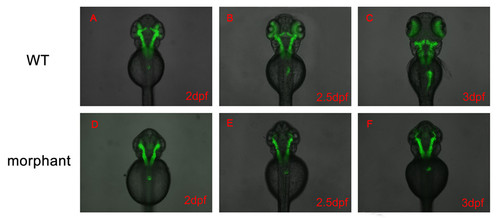
Nom1 affect exocrine pancreas development process between 2 dpf and 2.5 dpf. (A–F) All dorsal views, anterior to the top. (A, D) In morphant group, the GFP labeled exocrine pancreas size is the same as WT. (B, C, E, F) The exocrine pancreas began to enlarge in WT larvae, but not for the nom1-knockdown embryos at 2.5 dpf and 3 dpf. |