- Title
-
Super Resolution Microscopy Reveals that Caveolin-1 Is Required for Spatial Organization of CRFB1 and Subsequent Antiviral Signaling in Zebrafish
- Authors
- Gabor, K.A., Stevens, C.R., Pietraszewski, M.J., Gould, T.J., Shim, J., Yoder, J.A., Lam, S.H., Gong, Z., Hess, S.T., and Kim, C.H.
- Source
- Full text @ PLoS One
|
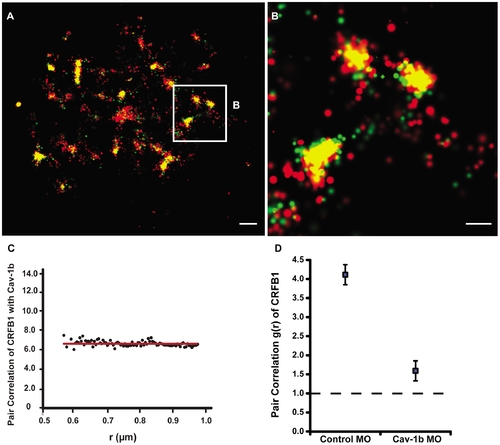
Cav-1b colocalizes with the zebrafish homolog of IFN-R and is positively correlated. ZFL cells (ne10) were transfected with Cav-1b-PAmCherry (red) and with CRFB1-dendra2 (green). For all images, 60×/1.2 NA magnification. Scale bars, 1 μm. Shown is the plasma membrane of one cell representative of the experiment (A) and a magnification (B) of the region marked by the white box in A. The image shows that Cav-1b and CRFB1 colocalize in the cell membrane. (C) Measurements of Cav-1b and CRFB1 show a positive pair correlation value g(r) greater than one, confirming that the two species are colocalized together. Pair correlation calculations were performed as described in Methods; briefly, g(r) >1 indicates positive correlation/clustering, and g(r) = 1 indicates a random distribution. (D) Pair correlation measurements of CRFB1 were calculated for the receptor, control morpholino (MO), and Cav-1b MO. CRFB1 is more prone to random distribution when Cav-1b is knocked down. Error bars SEM (n e8 cells). |
|
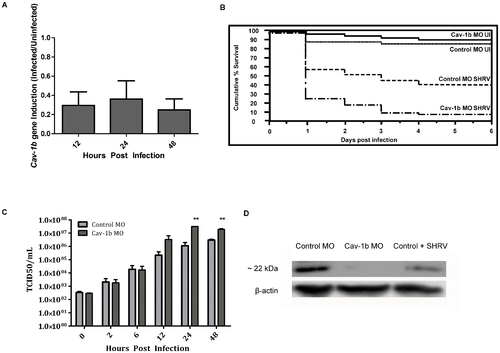
Cav-1b expression is modulated during virus infection, and Cav-1b knockdown leaves morphants susceptible to infection. A) Quantitative RT-PCR results revealed fold changes in the expression levels of Cav-1b in infected embryos when compared to uninfected embryos. Zebrafish were exposed seven dpf to 1×106 TCID50/mL virus. Total RNA was isolated from at 12, 24, and 48 hours post infection and reverse transcribed to cDNA (n = 20 fish per time point). All expression values have been normalized to the zebrafish β-actin gene. Error bars represent SEM of three replicates. B) Zebrafish embryos that were injected with Cav-1b morpholino (MO) to knock down the expression of Cav-1b or control MO were infected 48 hpf with 1×106 TCID50/ml virus and monitored for mortality. Results are representative of three separate experiments. Statistical analysis (Wilcoxon test) of the Kaplan-Meier curve was performed (*, p = 0.008). C) Zebrafish embryos that were injected with Cav-1b MO to knock down the expression of Cav-1b or control MO were infected by static immersion 48 hpf with 1×106 TCID50/ml virus. The graph indicates that early in infection (0–12 hpi), there is no difference in viral burden between Cav-1b morphants and controls. However, by 24–48 hpi, Cav-1b morphants have a higher viral burden. Figure is representative of three experiments; error bars are standard error of the mean (*, p<0.05). D) Western blot showing efficacy of MO knockdown in zebrafish. Zebrafish embryos from Control and Cav-1b MO, and Control MO with SHRV infection were compared for cav-1b expression at the 72 hpf developmental stage. At this time, infected fish were 24 hpi. Membranes were re-probed with antibody against β-actin to control for protein loading. PHENOTYPE:
|
|
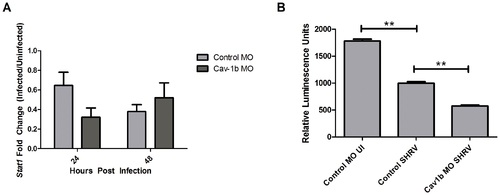
Decrease of Cav-1b expression negatively affects the IFN pathway. A) Stat1 gene expression was assessed by qRT-PCR in Control MO and Cav-1b MO embryos that were either SHRV infected or uninfected. Total RNA was extracted from 10 fish per treatment, cDNA synthesized and Stat1 mRNA expression assessed by qRT-PCR 24 hpi. The data are representative of three individual experiments and error bars indicate SEM. Each bar represents the mean fold induction of SHRV-infected embryos over corresponding controls. All expression values were normalized to zebrafish 18s. B) ISRE promoter activity is dampened in Cav-1b knockdown ZFL cells upon SHRV infection. ZFL cells were transfected with 250 ng of zISRE-luc construct along with 250 ng of cav-1b MO or control MO. Twenty four hours post transfection the ZFL cells were infected with SHRV at an MOI of 0.01. Cells were harvested for luciferase measurements 24 hpi. The graph shows relative luminescence units of control uninfected cells compared to cav-1b MO or control infected cells. Error bars are representative of SEM for two experiments. (**, p<0.001). |
|
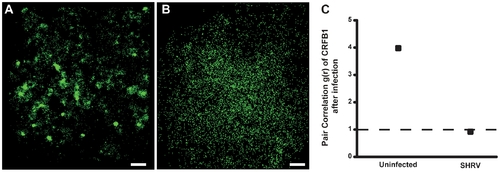
CRFB1 becomes dispersed as a result of whole virus infection in vitro. ZFL cells were infected 24 h post transfection and fixed prior to imaging. For all images, 60×/1.2 NA magnification. Scale bars, 1 μm. Shown for each part is the surface of one cell representative of the experiment. A) Uninfected cells overexpressing CRFB1 demonstrate that the receptor exists in clustered patches indicative of caveolae. B) Cells infected with SHRV demonstrate that CRFB1 becomes dispersed as a result of virus infection by 24 hpi. C) Pair correlation analysis confirms that compared to uninfected cells, CRFB1 becomes dispersed after infection. Values of g(r) in cells with SHRV infection are considered to be random in comparison to values in cells that remain uninfected (ne8 cells per treatment). |
|
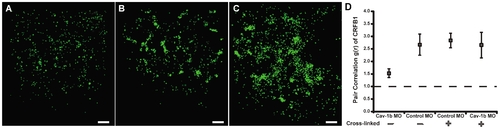
Crosslinking CRFB1 keeps receptor molecules clustered despite caveolin depletion. ZFL cells were co-transfected with MO and expression plasmid via nucleofection and allowed to recover/adhere to cell culture plates for ~6 hr prior to addition of crosslinking reagent. The crosslinking reaction was performed according to the manufacturer’s procedures. Cells were subsequently replenished with media and returned to the incubator for 24 hr post crosslinking. Scale bars, 1 μm. A) Cells transfected with Cav-1b MO/CRFB1 without crosslinking treatment show dispersed receptor molecules. B) Cells transfected with Control MO/CRFB1 with crosslinking clearly show clustered receptor molecules. C) Cells transfected with Cav-1b MO/CRFB1 with crosslinking. This demonstrates that despite depletion of Cav-1b, receptor molecules remain clustered. D) Pair correlation analysis confirms that with crosslinking, CRFB1 remains clustered despite Cav-1b depletion. Values of g(r) in cells with Cav-1b KD are similar to that for Controls (ne8 cells per treatment). |
|
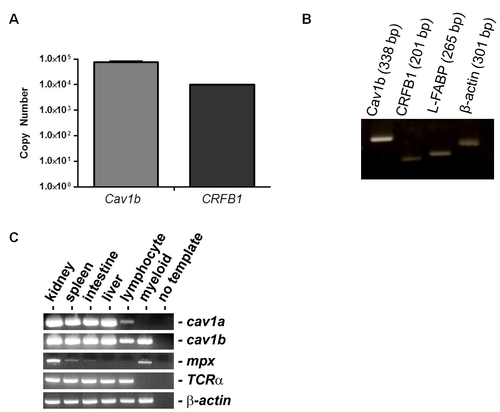
Caveolin-1 Expression in Cell Culture and Tissue-Specific Zebrafish cDNA Pools. A) qPCR demonstrates expression of endogenous cav1b and CRFB1 transcripts in RNA isolated from cultured ZFL cells. B) PCR was performed in liver tissue isolated from zebrafish embryos at the stage of virus infection (48 hpf) and demonstrates the expression of cav1b (338 bp), CRFB1 (201 bp), L-FABP (265 bp) and B-actin (301 bp) in the liver tissue of zebrafish embryos. C) PCR was performed to detect cav-1a and cav-1b gene expression in cDNA pools isolated from specific zebrafish tissues. Of note, cav-1b expression was detected in the kidney, lymphocyte, and myeloid lineages. |
|
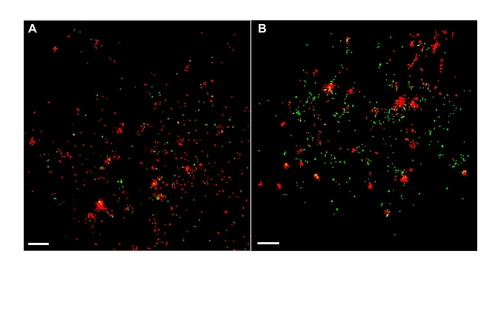
SHRV does not utilize caveolae to enter the host cell. FPALM imaging demonstrates no colocalization between fluorescently labeled virus and Cav-1b molecules early in the infection. Shown is a representative cell (total e8) of Cav-1b at 10 min post infection (A) and 2 h post infection (B). This indicates that SHRV does not use caveolae as a means of entry, suggesting that entry through caveolae will not be affected as a result of Cav-1 knockdown. For all images, 60×/1.2 NA. Scale bars, 1 μm. |
|
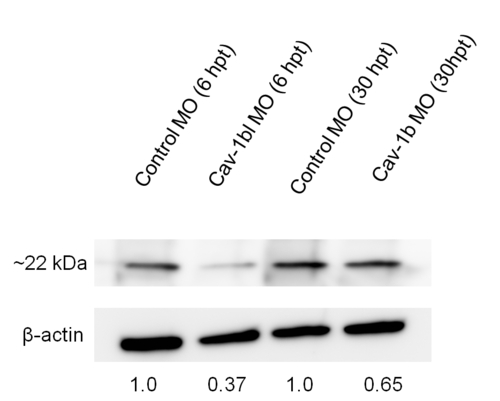
Knockdown of Cav-1b in ZFL cells. Western blot showing efficacy of MO (morpholino) in ZFL cells. Cells were transfected with Control morpholino (MO) or Cav-1b MO to knock down the expression of Cav-1b. Samples were taken at 6 and 30 h post transfection (hpt), corresponding to the time points used to perform experiments shown in Figure 7. At 6 hpt there is a marked decrease in cav-1 protein expression compared to control cells, while at 30 hpt a slight decrease in cav-1 protein expression is still observed. Membranes were re-probed with antibody against β-actin to control for protein loading. |