- Title
-
A cis-acting element in the coding region of cyclin B1 mRNA couples subcellular localization to translational timing
- Authors
- Yasuda, K., Kotani, T., and Yamashita, M.
- Source
- Full text @ Dev. Biol.
|
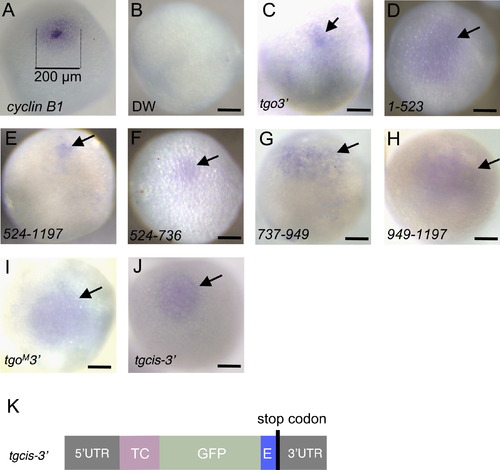
Localization of cyclin B1 reporter mRNAs that were transcribed from reporter genes injected into oocyte nuclei. (A) Whole-mount in situ hybridization of full-grown oocytes probed with cyclin B1. Aggregation of cyclin B1 mRNAs is found in the region indicated by dotted lines (200 μm). ((B)–(J)) Whole-mount in situ hybridization probed with gfp. Shown are oocytes injected with distilled water (DW) (B), reporter genes containing the full length (tgo3′) (C), 1–523 (D), 524–1197 (E), 524–736 (F), 737–949 (G) and 949–1197 (H) nts of the cyclin B1 coding region, tgoM3′ reporter gene (I) and tgcis-32 reporter gene (J). The oocytes injected with distilled water showed no signals (B). The tgo3′, 524–1197, 524–736 and tgcis–3′ mRNAs were aggregated in the animal polar cytoplasm ((C), (E), (F), (J)). In contrast, the 1–523, 737–949, 949–1197 and tgoM32 mRNAs were dispersed in the animal hemisphere of oocytes ((D), (G), (H), (I)). Arrows indicate the signals of reporter mRNAs. Bars, 100 µm. (K) Structure of the tgcis-3′ reporter gene, which contains cyclin B1 5′ UTR (52UTR), coding sequences of TC-tag (TC) and EGFP (GFP), CAGGAGACC element (E), a stop codon and cyclin B1 3′ UTR (3′UTR). |
|
Localization of cyclin B1 reporter mRNAs in oocytes of transgenic zebrafish. Whole-mount in situ hybridization of full-grown oocytes probed with cyclin B1 (A) or gfp ((B)–(I)). Shown are oocytes expressing tgo3′ (B), 1–523 (C), 524–1197 (D), 524–736 (E), 737–949 (F), 949–1197 (G), tgoM3′ (H) and SV40 (I) mRNAs. The tgo32, 524–1197 and 524–736 mRNAs were aggregated in the animal polar cytoplasm ((B), (D), (E)). In contrast, the 1–523, 737–949, 949–1197 and tgoM3′ mRNAs were dispersed in the animal hemisphere ((C), (F), (G), (H)) and the SV40 mRNAs were dispersed throughout the oocyte (I). Arrows indicate signals of cyclin B1 (A) and reporter mRNAs ((B)–(H)). Bars, 100 μm. (J) Structure of the SV40 reporter gene, which contains cyclin B1 5′ UTR (5′UTR), coding sequences of TC-tag (TC) and EGFP (GFP), a stop codon, cyclin B1 coding region and SV40 3′ UTR (SV40). (K) Quantification of reporter mRNAs (normalized to β-actin mRNA) in full-grown transgenic oocytes by real-time PCR. No statistically significant difference was found in the contents of reporter mRNAs. Error bars indicate mean±s.e.m. (n=3). |
|
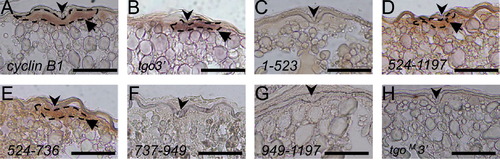
Localization of cyclin B1 reporter mRNAs in the animal polar cytoplasm of transgenic zebrafish oocytes. (A) Section in situ hybridization of a full-grown oocyte probed with cyclin B1. The cyclin B1 mRNAs were localized to the animal polar cytoplasm of the oocyte beneath the micro-pile as an aggregation (arrow). ((B)–(H)) Section in situ hybridization probed with gfp, showing full-grown oocytes of transgenic zebrafish expressing tgo3′ (B), 1–523 (C), 524–1197 (D), 524–736 (E), 737–949 (F), 949–1197 (G) and tgoM3′ (H) mRNAs. The tgo3′, 524–1197 and 524–736 mRNAs were aggregated beneath the micro-pile (arrows) ((B), (D), (E)), whereas the 1–523, 737–949, 949–1197 and tgoM3′ mRNAs had no signal ((C), (F), (G), (H)). Arrowheads indicate the micro-pile. Dotted lines encircle aggregated mRNAs. Bars, 100 μm. |
|
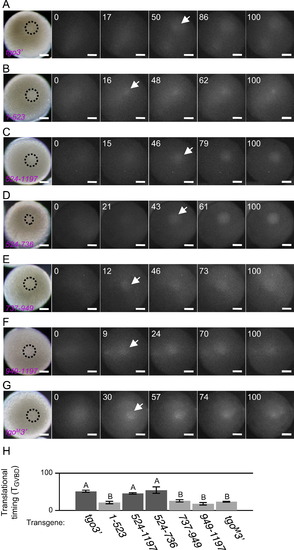
Real-time imaging of temporally regulated translation. ((A)–(G)) Real-time imaging of temporally regulated translation of tgo3′ (A), 1–523 (B), 524–1197 (C), 524–736 (D), 737–949 (E), 949–1197 (F) and tgoM32 mRNAs (G). The times after MIH stimulation are shown as standardized time TGVBD. Arrows indicate ReAsH signals detected at the first time after MIH stimulation. Dotted circles indicate the GV. Bars, 100 µm. (H) Translational timings of the reporter mRNAs after MIH stimulation. Error bars indicate mean±s.e.m. (n=3 for 1–523, 524–736 and tgoM32 n=5 for 949–1197; n=6 for 524–1197; n=7 for tgo3′ and 737–949). Transgenes indicated by B are translated earlier than those indicated by A, with statistically significant difference (P<0.05, Student2s t-test). |
|
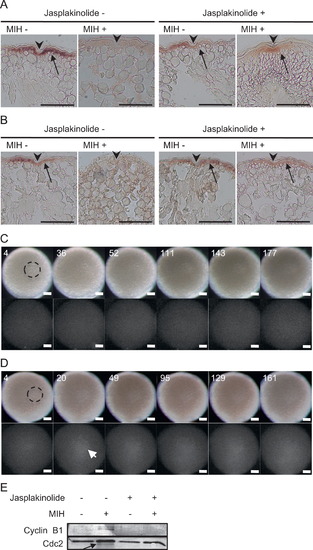
Effects of actin filament stabilization on cyclin B1 mRNA aggregation and translational regulation. ((A), (B)) Section in situ hybridization of wild-type oocytes probed with cyclin B1 (A) and tgo3′ mRNA-expressing oocytes probed with gfp (B). Oocytes were treated with jasplakinolide (+) or DMSO () and stimulated with (+) or without () MIH. The oocytes were fixed 140 min after MIH stimulation. Arrowheads indicate the micro-pile. Arrows indicate the signals of cyclin B1 mRNA (A) and tgo3′ reporter mRNA (B). ((C), (D)) Real-time imaging of oocytes expressing tgo3′ (C) or tgoM3′ mRNAs (D) treated with jasplakinolide and MIH. The times after MIH treatment (min) are shown. Bars, 100 μm. Dotted circles show the GV. Arrow indicates the initial translation signal. Similar results were obtained from six oocytes expressing tgo3′ mRNAs and three oocytes expressing tgoM3′ mRNAs. (E) Anti-Cyclin B1 and anti-Cdc2 immunoblots of oocytes treated with jasplakinolide (+) or DMSO (-) in the presence (+) or absence () of MIH. Cdc2 protein is a loading control of this experiment. An arrow indicates an active form of Cdc2 (Kondo et al., 2001). |
|
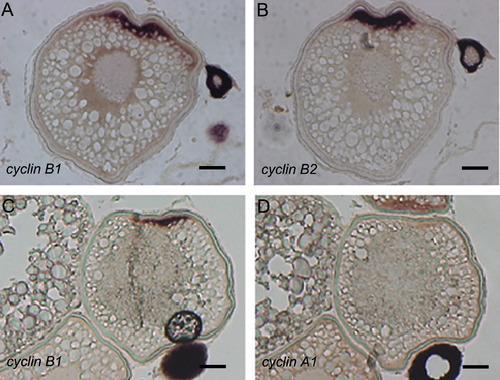
Localization of cyclin mRNAs in zebrafish oocytes. Serial sections ((A) and (B), (C) and (D)) of a full-grown oocyte were subjected to in situ hybridization analysis with cyclin B1 ((A), (C)), cyclin B2 (B) and cyclin A1 (D) probes. Note the co-localization of cyclin B1 and B2 mRNAs, but not cyclin A1 mRNA, to the animal polar cytoplasm. Bars, 100 μm. |
Reprinted from Developmental Biology, 382(2), Yasuda, K., Kotani, T., and Yamashita, M., A cis-acting element in the coding region of cyclin B1 mRNA couples subcellular localization to translational timing, 517-29, Copyright (2013) with permission from Elsevier. Full text @ Dev. Biol.