- Title
-
Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing
- Authors
- Obholzer, N., Swinburne, I.A., Schwab, E., Nechiporuk, A.V., Nicolson, T., and Megason, S.G.
- Source
- Full text @ Development
|
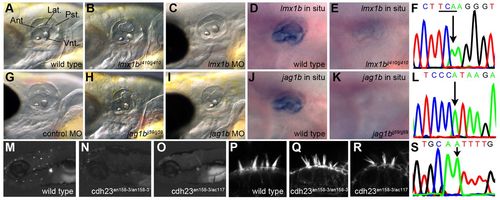
Validation of the identified ear mutants. (A) Lateral view of a wild-type ear at 65 hpf (fused projections labeled). Ant., anterior projection; Lat., lateral projection; Pst., posterior projection; Vnt., ventral projection. (B) Semicircular canal projections are abnormally shaped in jj410 ears at 65 hpf. (C) lmx1b morpholino knockdown recapitulates jj410 ear phenotype at 65 hpf. (D) lmx1b antisense in situ probe marks the inner ear and concentrates in semicircular canal projections. (E) Severe reduction of lmx1b transcript levels in a jj410 mutant larval inner ear at 65 hpf. (F) Sequencing trace of the jj410 genomic lesion in lmx1b; arrow denotes causative mutation that introduces a premature stop codon TGA (underlined, antisense strand was sequenced). (G) Control morpholino injection recapitulates wild type. (H) jj59 inner ears are shorter in the anterior-posterior axis and semicircular canal projections are malformed at 65 hpf. (I) jag1b morpholino knockdown recapitulates jj59 ear phenotype at 65 hpf. (J) jag1b antisense in situ probe marks the inner ear and concentrates in semicircular canal projections. (K) Severe reduction of jag1b transcript levels in a jj59 mutant larval inner ear at 65 hpf. (L) Sequencing trace of the jj59 genomic lesion in jag1b; arrow denotes causative mutation. (M) YO-PRO1 uptake by neuromast hair cells in wild-type larvae. (N) an158-3(cdh23nl9) mutant larval neuromast hair cells fail to take up YO-PRO1. (O) Failure to complement YO-PRO1 uptake in an158-3 x tc317e larval hair cells. (P) Intact inner ear hair cell bundles in the wild type. (Q) Hair cell bundles are splayed in an158-3(cdh23nl9) mutant inner ear hair cells. (R) Hair cell bundles show splayed phenotype and thus fails to complement in an158-3 x tc317e mutant inner ear. (S) Sequencing trace of the an158-3/cdh23nl9 genomic lesion in cdh23 (arrow). |
|
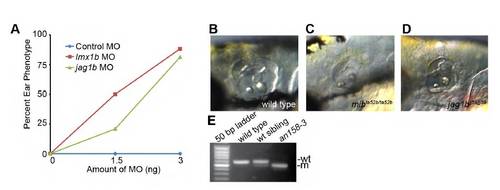
Additional validation of mutants supported their computed identities. (A) We observed a dose-dependent phenocopy of the mutant phenotypes with morpholinos against lmx1b and jag1b. (B-D) Compared with wild type (B), the ear phenotypes of mibta52b/ta52b (C) and jag1bjj59/jj59 (D) resemble one another and support the overlapping roles for both gene products in Notch signaling. (E) RT-PCR of cdh23 transcript in wild type, sibling and mutant reveals mis-splicing in an158-3(cdh23nl9) transcripts. PHENOTYPE:
|