- Title
-
An essential role of variant histone h3.3 for ectomesenchyme potential of the cranial neural crest
- Authors
- Cox, S.G., Kim, H., Garnett, A.T., Medeiros, D.M., An, W., and Crump, J.G.
- Source
- Full text @ PLoS Genet.
|
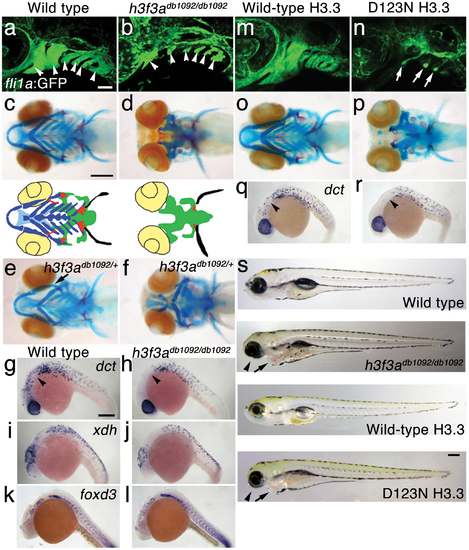
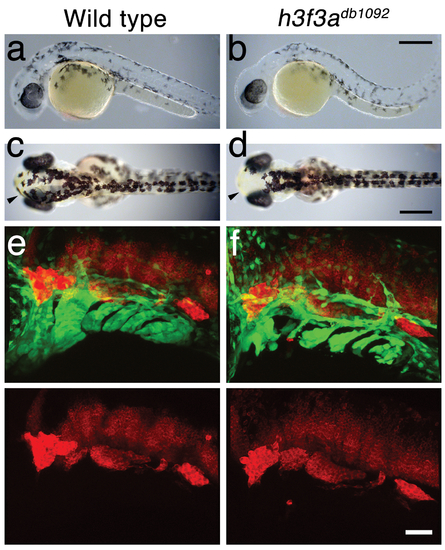
A dominant H3.3 mutation results in losses of CNC–derived head skeleton and pigment cells. a, b, fli1a:GFP-labeled arch ectomesenchyme (arrowheads) is greatly reduced, yet fli1a:GFP-positive endothelial cells (top) are unaffected, in both homozygous and heterozygous h3f3adb1092 mutants at 34 hpf (7/7 mutant; 0/10 wild-type). c, d, Homozygous h3f3adb1092/db1092 embryos specifically lack the CNC-derived head skeleton at 5 dpf (36/71 complete loss; 35/71 partial loss). Diagrams show the CNC-derived cartilage (blue) and bone and teeth (red), mesoderm-derived cartilage (green), pectoral fin cartilage (black), and eyes (yellow). e, f, h3f3adb1092/+ heterozygous larvae exhibit a wide range of craniofacial defects. In some cases, no defects are observed in the facial skeleton and heterozygotes are adult viable (not shown). In mild cases (e), dorsal cartilage and bone of the first and second arches are preferentially reduced, including the dorsal hyosymplectic cartilage and opercular bone of the second arch (arrow). In more severe cases (f), the cartilage and bone of the first arch and dorsal second arch are greatly reduced, with the anterior neurocranium and the posterior ceratobranchial cartilages being less affected. The frequency of skeletal phenotypes in h3f3adb1092/+ heterozygous larvae is highly variable between clutches. g, h, At 27 hpf, dct-positive melanophore precursors are selectively missing anterior to the ear (arrowheads) in both homozygous and heterozygous h3f3adb1092 embryos (5/5 mutant; 0/5 wild-type). i, j, At 27 hpf, cranial xdh-positive xanthophore precursors are mildly reduced in both homozygous and heterozygous h3f3adb1092 embryos (6/8 mutant; 0/4 wild-type). k, l, Wild-type and both homozygous and heterozygous h3f3adb1092 embryos have comparable numbers of foxd3-positive glial cells at 24 hpf (4 mutant; 5 wild-type). m-r, D123N h3f3a mRNA-injected but not wild-type h3f3a mRNA-injected embryos lack fli1a:GFP-positive ectomesenchyme (9/19 D123N; 0/12 wild-type), CNC-derived head skeleton (24/45 D123N; 0/21 wild-type) and cranial dct-positive melanophore precursors (anterior to the ear: arrowheads) (7/14 D123N; 0/11 wild-type). fli1a:GFP-positive blood vessels are unaffected (arrows). s, Except for the loss of the majority of the skull (no facial structures below the level of the eye: arrowheads) and mild heart edema (arrows), the overall morphologies of wild-type, homozygous and heterozygous h3f3adb1092, wild-type h3f3a-injected, and D123N h3f3a-injected larvae are indistinguishable at 5 dpf. Melanophores (black) and xanthophores (yellow) are also largely normal. Except for panels e and f, homozygous h3f3adb1092 examples are shown. Scale bars: a, b, m & n, 50 μm; c–l, o–s, 250 μm. |
|
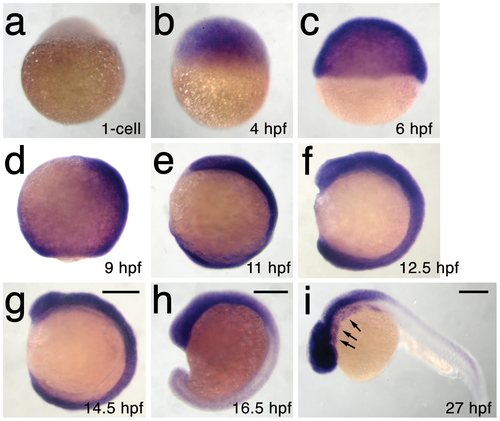
h3f3a is ubiquitously expressed throughout embryogenesis. a–i, Lack of h3f3a expression at the one-cell stage shows that h3f3a mRNA is not maternally provided. From 4–14.5 hpf, h3f3a is expressed ubiquitously throughout the embryo. By 16.5 hpf and 27 hpf, h3f3a expression is still widespread, with higher levels apparent in the anterior part of the embryo, including the CNC-derived ectomesenchyme of the pharyngeal arches (arrows) at 27 hpf. Scale bars = 250 μm. |
|
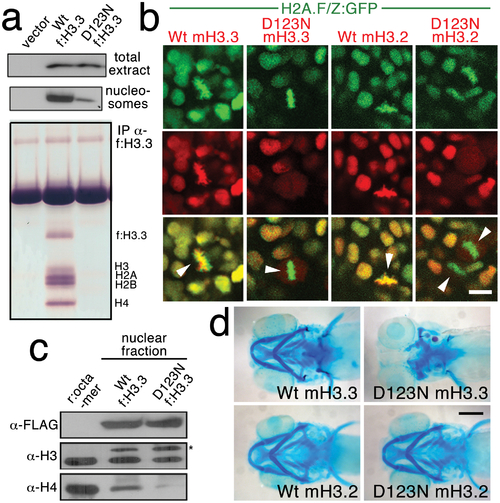
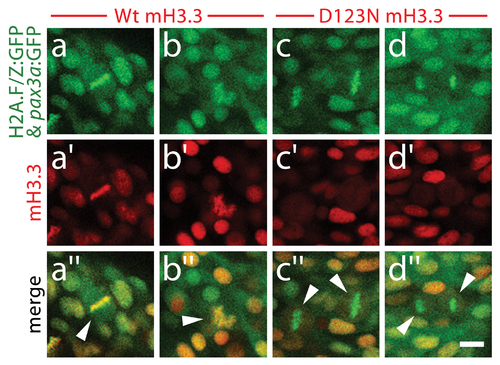
The dominant D123N mutation prevents chromatin incorporation and promotes the formation of aberrant H3 homodimers. a, Western blots show α-FLAG immunostaining of nuclear extracts or purified mononucleosome fractions from HEK cells transfected with vector alone or FLAG-tagged H3.3 (f:H3.3) vectors. Wild-type and D123N f:H3.3 proteins are expressed at equal levels in total extract, but wild-type f:H3.3 is present at much higher levels in the nucleosome fraction (consistent over three replicate experiments). α-FLAG immunoprecipitation from purified nucleosomes shows that wild-type but not D123N f:H3.3 is incorporated into nucleosomes containing H2A, H2B, H3, and H4 (consistent over three replicate experiments). b, Confocal images from H2A.F/Z:GFP embryos expressing wild-type and D123N versions of mCherry(m)H3.3 and mCherry(m)H3.2 fusion proteins. Merged images show that whereas all H3 proteins are nuclear localized in surrounding non-mitotic cells, wild-type mH3.3 and mH3.2, but not D123N mH3.3 and mH3.2, co-localize with H2A.F/Z:GFP in the chromosomes of metaphase/anaphase cells (arrowheads) after nuclear envelope breakdown (wild-type mH3.3, 11/11 cells in 2 embryos; D123N mH3.3, 0/25 cells in 3 embryos; wild-type mH3.2, 21/21 cells in 3 embryos; D123N mH3.2, 0/16 cells in 2 embryos). c, α-FLAG, α-H3 and α-H4 western blots for samples immunoprecipitated by α-FLAG from nuclear extracts of f:H3.3-transfected HEK cells. Recombinant octamer is used as a reference. Whereas both endogenous H3 and H4 co-immunoprecipitate with the wild-type f:H3.3 protein (*), H3 but not H4 co-immunoprecipitates with D123N f:H3.3 (asterisk marks the larger recombinant f:H3.3 protein). Results were consistent over three replicate experiments. d, mRNA injection of D123N mH3.3 (8/17), but not wild-type mH3.3 (0/26), wild-type mH3.2 (0/19), or D123N mH3.2 (0/18), results in loss of the CNC-derived head skeleton at 4 dpf. Scale bars: b, 10 μm; d, 250 μm. |
|
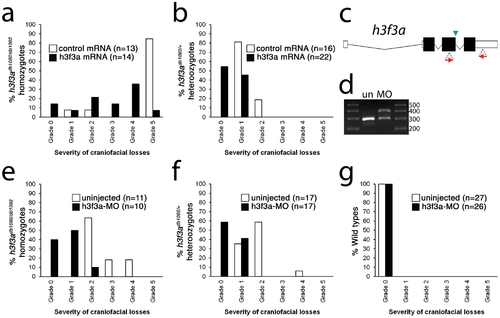
Injection of wild-type H3.3 RNA and reduction of mutant H3.3 levels both rescue craniofacial skeletal development in h3f3adb1092 mutants. a, b, Results from scoring the severity of craniofacial losses in 5 dpf larval head skeletons from h3f3adb1092 siblings injected with 450 ng/µl mRNA encoding wild-type H3.3 or a control kikGR fluorescent protein. The scoring system ranges from Grade 0 (wild-type phenotype) to Grade 5 (complete loss of CNC derivatives); see Materials and Methods for more detail. H3.3-mRNA-injected h3f3adb1092 homozygotes (a) and heterozygotes (b) exhibited a decrease in the severity of h3f3adb1092 craniofacial phenotypes over kikGR-RNA-injected controls (significant by Fisher′s exact test: homozygotes, p = 7.7E-05; heterozygotes, p = 1.0E-04). c, An antisense morpholino oligonucleotide was designed to inhibit splicing at the exon 3/intron 3–4 boundary (green arrowhead) of the h3f3a transcript. Morpholinos were injected into one-cell-stage h3f3adb1092 embryos at 400 µM. d, Morpholino efficacy was demonstrated by PCR amplification between exons flanking the targeted splice junction from 10 hpf cDNA from 20 pooled embryos (position of primers shown as red arrows in c). Compared to the sample from uninjected (un) embryos, the morpholino-treated sample (MO) exhibited a partial decrease in PCR product representing spliced transcript (295 bp) and a concomitant increase in un-spliced PCR product (390 bp). e, f, Compared to uninjected siblings, both morpholino-injected h3f3adb1092 homozygotes (e) and heterozygotes (f) exhibited a decrease in the severity of h3f3adb1092 craniofacial phenotypes (significant by Fisher′s exact test: homozygotes, p = 2.0E-04; heterozygotes, p = 6.3E-06). (g) Craniofacial development in wild-type embryos was unaffected by morpholino injection. |
|
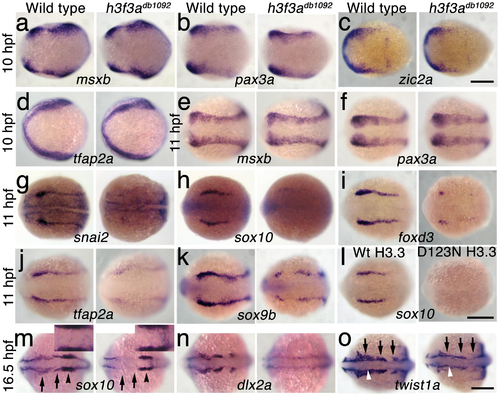
H3.3 functions at the NPB–CNC transition. a–f, Expression of msxb, pax3a, zic2a, and tfap2a at 10 hpf and msxb and pax3a at 11 hpf is indistinguishable between wild types and both homozygous and heterozygous h3f3adb1092 mutants (mut) (n≥10 for each). g–k, At 11 hpf, both homozygous and heterozygous h3f3adb1092 mutants have severe reductions in the expression of snai2 (4/4 mut; 0/4 wt), sox10 (10/12 mut; 0/6 wild-type), foxd3 (9/9 mut; 0/5 wt), tfap2a (7/7 mut; 0/4 wt), and sox9b (8/8 mut; 0/3 wt). l, sox10 expression is also lost in embryos injected with D123N (10/12) but not wild-type (0/16) h3f3a mRNA. m, In both homozygous and heterozygous h3f3adb1092 embryos, sox10 expression partially recovers by 16.5 hpf yet is specifically reduced in presumptive CNC ectomesenchyme domains (arrows) (6/6 mut; 0/4 wt). An increase in sox10-positive cells is evident in the mutant dorsal neural tube (insert) between the sox10-positive otic placodes (arrowheads) which are unaffected in mutants. n, At 16.5 hpf, dlx2a expression in three streams of migrating ectomesenchyme is reduced in both homozygous and heterozygous h3f3adb1092 mutants (6/7 mut; 0/5 wt). o, The 16.5 hpf ectomesenchyme expression (arrows) of twist1a is reduced in both homozygous and heterozygous h3f3adb1092 mutants yet paraxial mesoderm expression is unaffected (white arrowheads) (8/8 mut; 0/5 wt). In all panels, homozygous h3f3adb1092 examples are shown. All images are dorsal views with anterior to the left. Scale bars: 250 μm. EXPRESSION / LABELING:
|
|
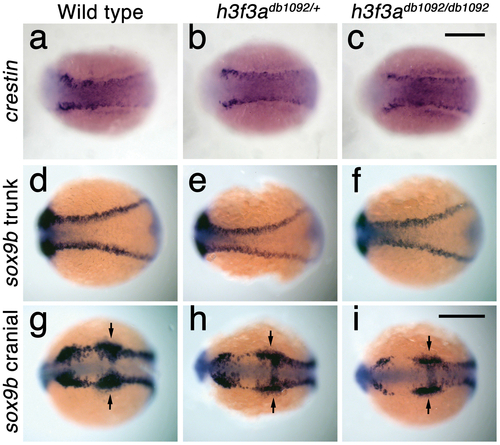
Trunk NC is largely unaffected in h3f3adb1092 mutants. a–c, crestin expression at 11.7 hpf shows similar amounts of trunk NC in wild-type and both homozygous and heterozygous h3f3adb1092 embryos (n = 5 for each genotype). d–i, Trunk views of sox9b expression at 11.7 hpf show that trunk NC specification is largely normal in both homozygous and heterozygous h3f3adb1092 mutants (n = 10 for each genotype). Cranial views of the same embryos show reduced amounts of sox9b-expressing CNC. Arrows show the sox9b-positive otic placodes that are unaffected in mutants. Scale bars = 250 μm. EXPRESSION / LABELING:
|
|
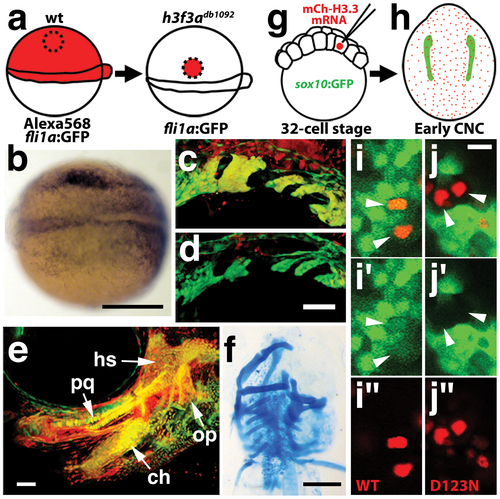
H3.3 function is required tissue- and cell-autonomously for CNC development. a, Wild-type cells were transplanted unilaterally into the CNC precursor domain of h3f3adb1092/db1092 homozygous mutants at 6 hpf. b, Compared to the non-recipient control side (bottom), expression of the early CNC marker snai2 is restored in the recipient side at 11hpf (top) (n = 17/29 with rescue). c–e, fli1a:GFP (green) marks CNC ectomesenchyme of the pharyngeal arches at 30 hpf and facial skeletal elements at 5 dpf. The red fluorescent dye, Alexa568, marks transplanted wild-type cells, whereas both donor and host cells harbor the fli1a:GFP transgene. When transplanted into an h3f3adb1092/db1092 homozygous host, wild-type Alexa568+ CNC precursors contribute to pharyngeal arch ectomesenchyme and rescue arch size (c) and form wild-type cartilage and bone (e). In contrast, the non-recipient control side (d) has reduced pharyngeal arch ectomesenchyme. Whereas wild-type donor cells appear yellow due to red Alexa568 and green fli1a:GFP, mutant host cells have only fli1a:GFP and hence appear green. Rescue of arch size was observed in 8/11 cases. f, Alcian staining at 5 dpf shows that cartilage is restored to half the face in an h3f3adb1092/db1092 larvae that received an unilateral wild-type CNC precursor transplant. Compare the recipient side (left) to the control side that forms little facial cartilage (right). Skeletal rescue was observed in 21/30 cases. g, Individual cells of 32-cell stage sox10:GFP embryos were injected with mRNA encoding mCherry-tagged versions of wild-type or D123N H3.3 to generate mosaic mCherry-H3.3 expression at later stages. h, Soon after the appearance of GFP-labeled CNC at approximately 11 hpf, mosaic embryos were assessed for incorporation of mCherry-H3.3-expressing cells (red) into the sox10:GFP-positive CNC domain (green). i/i′/i′′ and j/j′/j′′, Confocal images from sox10:GFP embryos with mosaic expression of wild-type (i/i′/i′′) and D123N (j/j′/j′′) versions of mCherry-H3.3. Cells doubly-positive for wild-type mCherry-H3.3 (red) and GFP (green) (arrowheads) were observed within the CNC domain (7/14 cells over 4 embryos), whereas mutant D123N mCherry-H3.3 cells within the CNC domain failed to up-regulate sox10:GFP (arrowheads) (0/18 cells over 4 embryos). Only cells with strong mCherry-H3.3 were used in the analysis. hs: hyosymplectic cartilage, pq: palatoquadrate cartilage, ch: ceratohyal cartilage, op: opercular bone. Scale bars: b, f, 250 μm; c–e, 50 μm; i and j,10 μm. |
|
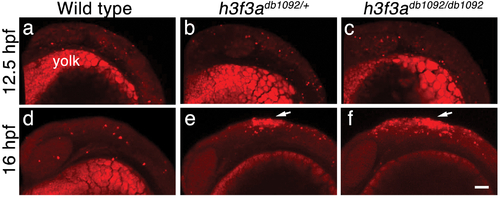
Cell death in h3f3adb1092 embryos. a–c, Lysotracker Red staining marks similar amounts of dying cells in wild-type (n = 2), h3f3adb1092/+ heterozygous (n = 5), and h3f3adb1092/db1092 homozygous (n = 3) embryos at 12.5 hpf. The bright staining in the bottom of each panel is the yolk. d–f, At 16 hpf, increased Lysotracker Red staining (arrows) was evident in the dorsal neural tube of h3f3adb1092/+ heterozygotes (2/2) and h3f3adb1092/db1092 homozygotes (3/3) but not wild types (0/4). These dying cells were located in a similar position to where CNC forms in wild-type embryos. Scale bar = 50 μm. PHENOTYPE:
|
|
NC derivatives in h3f3adb1092 mutants. a–d, Live views at 32 hpf (a, b) and 54 hpf (c, d) show reductions of head melanocytes (black) in h3f3adb1092/db1092 homozygotes (n = 22/28). The melanocytes of the eye and trunk were never affected. Cranial xanthophores (yellow), most clearly seen at the anterior limit of the head (arrowheads), were also largely unaffected. e, f, In confocal projections of fli1a:GFP embryos at 36 hpf, HuC antibody staining (red) labels neurons of the cranial ganglia – from left to right the trigeminal, anterior lateral line, auditory, and posterior lateral line – which are unaffected in h3f3adb1092/db1092 homozygotes (n = 8). In the merged images, fli1a:GFP (green) shows a reduction of CNC-derived ectomesenchyme in the mutant. Scale bars: a–d, 250 μm; e & f, 50 μm. |
|
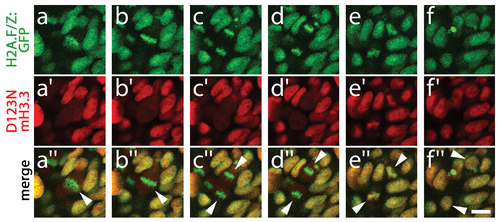
D123N H3.3 fails to localize to condensed chromosomes within NPB cells. Confocal images of 13 hpf embryos harboring the H2A.F/Z:GFP and NPB-specific pax3a:GFP transgenes and injected with wild-type or D123N versions of mCherry(m)H3.3 fusion proteins. a–d, GFP fluorescence of cells within the pax3a:GFP-labeled NPB domain. Whereas most cells are in interphase, the few cells in metaphase/anaphase (arrowheads in merged images: a′′–d′′) exhibit both GFP-labeled condensed chromosomes (H2A.F/Z:GFP) and more diffuse lower-level cytoplasmic GFP (pax3a:GFP). a2–b2, Wild-type mCherry-H3.3 localizes within the condensed chromosomes of 14/14 metaphase/anaphase cells. c′–d′, D123N mCherry-H3.3 fails to localize within condensed chromosomes and instead appears diffuse throughout pax3a:GFP-positive NPB cells after nuclear envelope breakdown. (0/14 cells exhibit chromosomal localization during metaphase/anaphase). Scale bar = 10 μm. |
|
D123N H3.3 protein remains stable during mitosis. Time course of confocal images from H2A.F/Z:GFP embryos expressing D123N mCherry(m)H3.3 fusion protein, showing a cell (arrowhead in merged image a′′) progressing through the stages of mitosis including metaphase (a/a′/a′′ and b/b′/b′′), anaphase (c/c′/c′′ and d/d′/d′′), telophase (e/e′/e′′) and the eventual establishment of two new daughter cells (arrowheads, f/f′/f′′). a–d, H2A.F/Z:GFP localizes to condensed chromosomes during both metaphase and anaphase. a2–d2, In contrast, D123N mCherry-H3.3 fails to co-localize with H2A.F/Z:GFP and appears as a weak diffuse signal throughout the cell(s) after nuclear envelope breakdown. e/e′/e′′, H2A.F/Z:GFP and D123N mCherry-H3.3 subsequently become co-localized during the re-establishment of the nuclear membranes during telophase. f/f′/f′′, Nuclear co-localization continues into interphase in both daughter cells. The rapid re-appearance of strong nuclear mCherry-H3.3 signal in telophase (16/16 cells over 2 embryos) confirms that the low-level diffuse D123N mCherry-H3.3 signal observed during metaphase/anaphase results from a failure to localize to condensed chromosomes rather than protein degradation. Scale bar = 10 μm. |
|
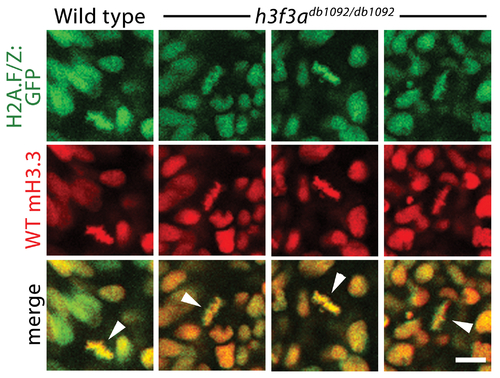
Localization of wild-type mCherry-H3.3 within metaphase/anaphase cells of h3f3adb1092/db1092 embryos. Confocal images from wild-type and h3f3adb1092/db1092 homozygous embryos harboring the H2A.F/Z:GFP transgene and injected with mRNA encoding wild-type mCherry(m)H3.3 fusion protein. Merged images show that wild-type mH3.3 protein co-localizes with H2A.F/Z:GFP in the chromosomes of metaphase/anaphase cells (metaphase cells shown: arrowheads) in wild-type and h3f3adb1092/db1092 homozygotes (mutant, 21/21 cells in 3 embryos; wild-type, 15/15 cells in 3 embryos). Detailed analysis of fluorescence levels revealed no significant differences in the distribution of wild-type mH3.3 fluorescence between wild types and mutants. Scale bar = 10 μm. |
|
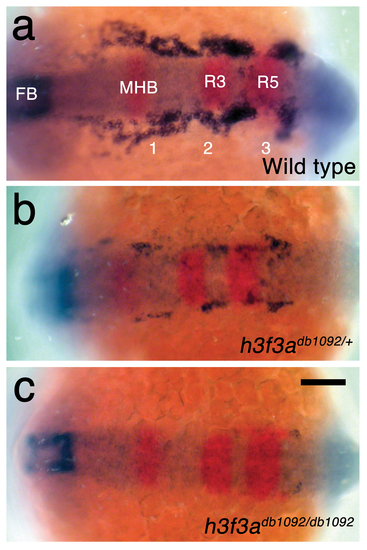
h3f3adb1092 embryos lack CNC ectomesenchyme but have normal neural patterning. a, In wild-type embryos at 15.5 hpf, expression of dlx2a (blue) marks a subset of the forebrain (FB) and three streams of migrating CNC-derived ectomesenchyme (1–3). In red, pax2a expression marks the mid-hindbrain boundary (MHB) and egr2b expression marks rhombomeres 3 and 5 (R3 and R5) of the hindbrain. b, c, dlx2a-positive ectomesenchyme is reduced (n = 18/18) in h3f3adb1092/+ heterozygous embryos and completely lost (n = 3/9) or greatly reduced (n = 6/9) in h3f3adb1092 homozygotes at similar stages. Neural patterning was never affected in h3f3adb1092 heterozygotes and homozygotes. Scale bar = 100 μm. |
|
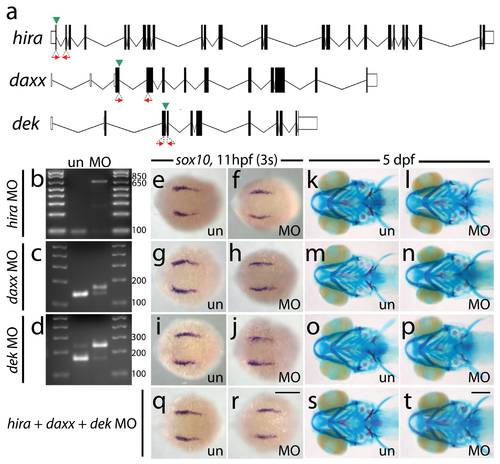
Antisense morpholino targeting of H3.3 chaperones. a, Zebrafish have one predicted copy each of hira, daxx, and dek genes in their genomes. Antisense morpholino oligonucleotides were designed to inhibit splicing at specific exon/intron boundaries (green arrowheads) within hira (exon 1/intron 1–2), daxx (exon 3/intron 3–4), and dek (exon 3/intron 3–4) transcripts. Morpholinos were injected into one-cell-stage zebrafish embryos at 400 μM. b–d, Morpholino efficacy was demonstrated by PCR amplification between exons flanking targeted splice junctions from 10 hpf cDNA from 20 pooled embryos (position of primers shown as red arrows in a). Morpholino-treated samples exhibited a significant decrease in PCR product representing spliced transcript (b, hira, 96 bp; c, daxx, 143 bp; d, dek, 169 bp) and a concomitant increase in un-spliced PCR product (b, hira, 637 bp; d, dek, 247 bp) or an alternative spliced transcript (c, daxx, 169 bp: utilization of cryptic splice donor site 26 nucleotides into adjacent intron, predicted to result in frameshift and early termination). e–j, Wholemount in situ hybridization for sox10 at 11 hpf. Morpholinos against hira (f), daxx (h) and dek (j) have no effect on early sox10 expression within CNC cells when compared to uninjected controls (e, g, i) (n≥9 for each). k–p, 5 dpf larval head skeletons stained with Alcian blue (cartilage) and Alizarin red (bone and teeth). Craniofacial development is unaffected in surviving hira (l), daxx (n) and dek (p) morpholino-treated individuals and uninjected controls (k, m, o) (n≥35 for each). hira morpholino-injected embryos did exhibit a high level of death after 24 hpf but prior to 5 dpf (morpholino injected, 55.1%; uninjected, 0%) and a curved/kinked tail phenotype in surviving 5 dpf larvae (morpholino injected, 48.6%; uninjected, 0%). q–t, Compared to uninjected controls (q & s), embryos injected with a combination of all three morpholinos at 200 μM had no defects in CNC sox10 expression (r) (n≥9 for each) or development of the larval head skeleton (t) (n≥66 for each). un, uninjected; MO, morpholino. Scale bars = 250 μm. |