- Title
-
Maternal pak4 Expression is Required for Primitive Myelopoiesis in Zebrafish
- Authors
- Law, S.H., and Sargent, T.D.
- Source
- Full text @ Mech. Dev.
|
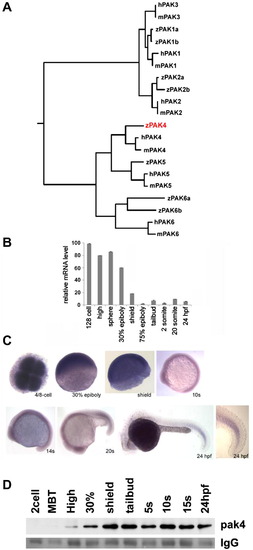
Identification and expression of pak4 in zebrafish embryos. (A) Phylogeny of the pak family. Putative protein sequences of the kinase domains of human (h), mouse (m) and zebrafish (z) PAKs were analyzed using ClustalW (http://clustalw.genome.jp/). Results are shown as a dendrogram with branch length indicating divergence. PAK5 proteins were also named as PAK7 in Genbank and EnsEMBL databases. Genbank Accessions or EnsEMBL Gene IDs (Zv9): humanPAK1, NM_001128620; humanPAK2, NM_002577; humanPAK3, NM_001128166; humanPAK4, NM_005884; humanPAK5, NM_020341; humanPAK6, NM_020168; mousePAK1, NM_011035; mousePAK2, NM_177326; mousePAK3, NM_008778; mousePAK4, NM_027470; mousePAK5, NM_172858; mousePAK6, NM_001033254; zfPAK1a, ENSDARG00000042624; zPAK1b, NM_201328; zPAK2a, NM_001002717; zPAK2b, NM_001025456; zPAK4, NM_001002222; zPAK5, NM_212962; zfPAK6a, ENSDARG00000027564; zPAK6b, NM_001162487. Zebrafish PAK4 is indicated in red. (B) Quantitative analysis of zebrafish pak4 expression in development. Quantitative RT-PCR was carried out with total RNA (800 ng) extracted from specific embryonic stages and analyzed for pak4 expression. Maternal pak4 mRNA levels rapidly declined between 30% epiboly and shield stages, then remained approximately constant through 24 hpf. Results shown were collected from three individual experiments. (C) Whole-mount in situ hybridization for pak4. Stages are as indicated. 4/8-cell stage – animal pole view, others are lateral view. Signals at and after 30% epiboly were relatively weak and hence staining of these samples for alkaline phosphatase activity was prolonged. A higher magnification of the tail of the 24 hpf embryo is shown to highlight elevated expression in newly formed somites. (D) Immunoprecipitation/western analysis. Total soluble protein was isolated from embryos at multiple stages, immunoprecipitated and prepared for western blotting as described in Section 2. Equal numbers of embryos were used for each sample, which was confirmed by BCA protein assays. Ponceau S staining of the IgG heavy chain band (IgG) is shown below as a control for immunoprecipitation recovery. |
|
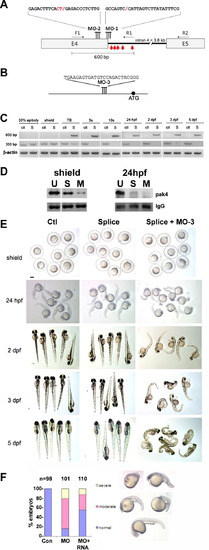
Loss of function analysis. (A) Splice-blocking MO design. The sequences of MO-1 and MO-2 and their target sites at the 32-end of pak4 exon 4 are shown. Two MOs were necessary for complete knockdown due to the unmasking of a cryptic splice donor. F1, R1 and R2 refer to PCR primers used to monitor splicing (see Section 2). The red arrows indicate the positions of in-frame stop codons in the abortively spliced mRNA. (B) Translation-blocking MO of the indicated sequence targeting a site upstream from the start codon (black circle). (C) RT-PCR analysis of splice-blocking. Embryonic RNA from the indicated stages was isolated after injection of the control MO (ctl) or combined splice-blocking MOs (S) and used as template for PCR. The 300-bp PCR product was amplified by the primer pair F1 + R2 and represents the correctly spliced pak4 mRNA, while the 600-bp product (primers F1 + R1) represents the abortively spliced mRNA. The 600-bp product appeared at tailbud stage (TB), indicating pak4 transcription from the zygotic genome. At this stage the 300-bp product had declined in splice-inhibited embryos and had disappeared by the 5-somite stage (5s). This pattern persisted through the final stage tested, 5 dpf. As a positive control a fragment of β-actin was amplified in the corresponding samples. (D) MO effectiveness. Pak4 protein was detected by immunoprecipitation/western blotting from equal numbers of embryos injected with the splice MOs (S; MO-1, MO-2) or a mixture of splice- and translation-blocking MOs (M; MO-3), or from uninjected embryos (U). Equal protein input for immunoprecipitations was confirmed by BCA assays. At shield stage the splice MOs reduced pak4 partially, while the splice + translation-blocking mixture had a much stronger effect. Both treatments gave essentially complete inhibition at 24 hpf, although some residual pak4 protein was detected in the splice morphants. Ponceau S staining of the IgG heavy chain band (IgG) is shown below as a control for immunoprecipitation recovery. (E) Representative embryos at various stages injected with control (Ctl), splice-blocking MOs (Splice; 1.5 + 1.5 ng), or a combination of the translation- (3 ng) and splice-blocking (1.5 + 1.5 ng) MOs (Splice + MO-3). Only the latter showed disrupted morphology, as observed at 24 hpf and steadily worsening through 5 dpf. Scale bar, 250 μM. (F) Summary of pak4 mRNA rescue data at 24 hpf. Morphant embryos were classified as normal, moderate (bent axis, small head and eyes), or severe (kinked and shortened axis, small head and eyes, unidentified forebrain and hindbrain, or missing head and tail – “monster”). The proportion and severity of affected embryos were both decreased substantially by co-injection of 900 pg pak4 mRNA. |
|
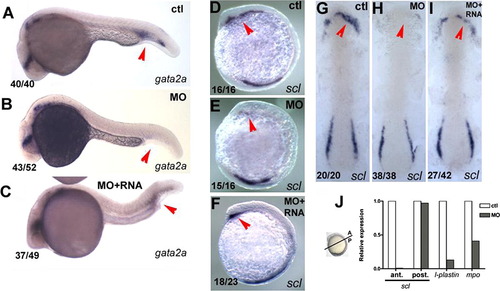
pak4 is required for primitive myelopoiesis. (A–C) Inhibition of gata2a expression by MZpak4 knockdown in the posterior blood island (red arrows) in 24-hpf embryos. (D–I) Inhibited expression of the hematopoietic regulatory factor scl in the rostral blood island (RBI; red arrows) at 5-somite stage, lateral view (D–F) and 7-somite stage flat mounts (G–I). (A, D and G) Control morphants, (B, E and H) MZpak4 knockdown embryos. (C, F and I) MZpak4 knockdown embryos rescued by pak4 mRNA injection. (J) Relative expression levels of scl in anterior and posterior regions at 5-somite stage, l-plastin at 15-somite and mpo at 20-somite stages. mRNA levels were measured by QPCR, normalized to 18S rRNA levels and represented as a ratio to the corresponding uninjected control embryos. Samples for anterior and posterior measurement of scl were dissected across the axis as shown while those for l-plastin and mpo were from whole embryos. Results shown are representatives from two individual experiments with similar results. EXPRESSION / LABELING:
PHENOTYPE:
|
|
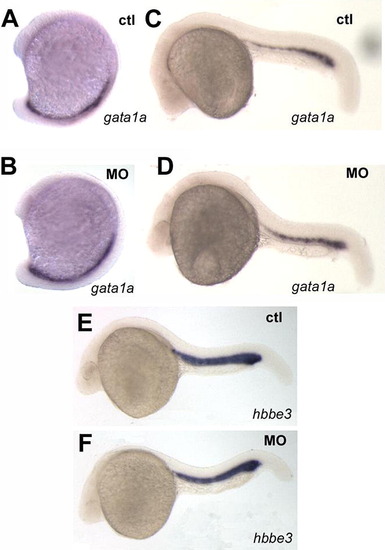
Primitive erythropoiesis is not affected by pak4 knockdown. Two specific markers for erythroid precursors, gata1a and hbbe3 in the posterior blood lineage and intermediate cell mass were unaffected by pak4 MO injection at 15-somite (A and B) or 24 hpf (C–F). EXPRESSION / LABELING:
PHENOTYPE:
|
|
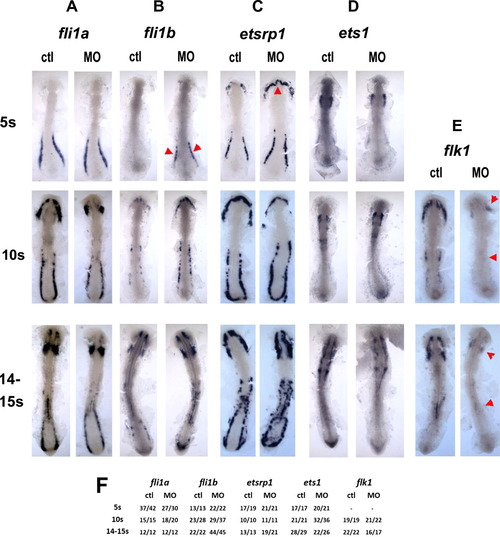
Regulatory level of pak4 function. Flat mounts of in situ hybridization for ETS class transcription factors fli1a (column A) and fli1b (column B), etsrp1 (column C) and ets1 (column D), and the VEGF receptor flk1 (column E) reveal different dependence on pak4. Expression of the ETS transcription factors was not inhibited at any stage tested, however, expression of fli1b was transiently elevated at in the posterior domain, and etsrp1 in the rostral domain at the 5-somite stage (B and C; red arrowheads). flk1 Expression was reduced in both the anterior and posterior hematopoietic domains (E; red arrowheads). Frequency data are tabulated in (F). EXPRESSION / LABELING:
|
|
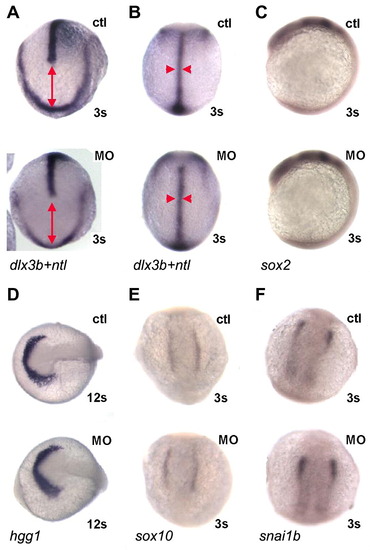
MZpak4 knockdown did not disturb early signaling pathways. (A and B) Convergent extension: double in situ hybridization with dlx3b and ntl probes was performed to outline the neural plate border and notochord at 3-somite stage. Dorsal view of anterior (A) and trunk (B) regions. The distance between dlx3b and the leading edge of ntl expression indicates extension (red line in A) while the width of ntl (red arrowheads in B) indicates convergence. The patterns in MZpak4 knockdown (bottom) are comparable to control embryos (top), indicating normal convergent extension in gastrulation. (C–F) Early markers for neural plate (sox2; C), hatching gland (hgg1; D) and neural crest (sox10; E and snai1b; F) were unaffected in pattern or intensity by MZpak4 knockdown (bottom embryos) compared to controls (top). Embryos were 12 somite stage in panel D, 3 somite stage in the other panels. PHENOTYPE:
|
|
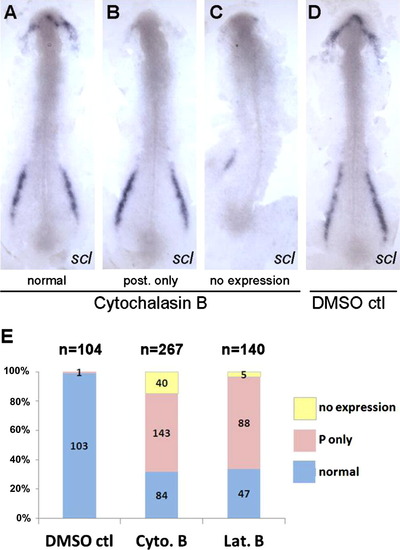
Disruption of actin microfilaments leads to loss of scl expression in the RBI. Dechorinated embryos at tailbud stage were exposed to cytochalasin B (4 μg/ml) or latrunculin B (0.25 μg/ml) and fixed at 5–6 somite stage for whole-mount in situ hybridization using the ,scl gene probe. 30–40% Mortality was observed in all drug exposures due to rupture of the enveloping layer at variable times. Dead embryos were not analyzed further. Flat mounts of representative embryos from cytochalasin B exposure (A–C; normal, posterior expression only, no expression) and DMSO control (D) were prepared to show the anterior and posterior expression of scl. (E) Data were summarized from four independent experiments which show that the majority of embryos specifically lost the anterior scl expression after drug treatments. Posterior-specific loss of expression was never observed. |
|
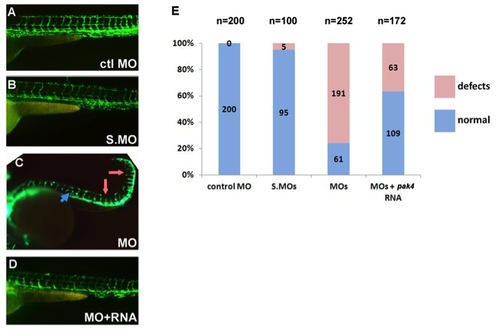
Vascular defects in MZpak4 morphants. fli1a:eGFP transgenic embryos were injected with control MO (A), splice-blocking MOs (B), or the combination of the translation- (MO-3) and splice-blocking MOs (C). As indicated by the fluorescing endothelial cells, splice morphants exhibited normal vasculature (B), while defects were observed in the MZpak4 knockdown embryos (C), including short intersegmental vessels (ISV; pink arrows), abnormal longitudinal extension of ISV at myoseptum (blue arrow) and partial loss of dorsal longitudinal anastomotic vessels (DLAV). The defects could be substantially rescued by co-injection of pak4 mRNA (D). Combined results from three experiments are summarized by the histogram in (E). Images were recorded at 50 hpf. |
|
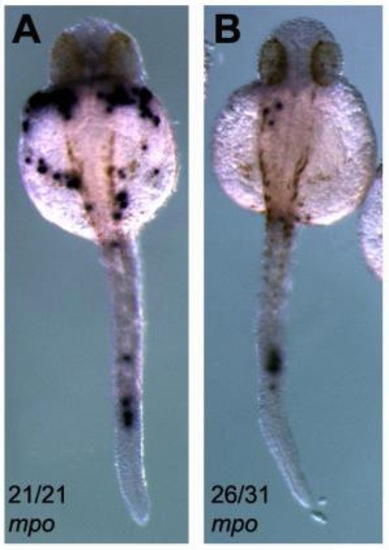
Expression of mpo in MZpak4 morphants. At 30 hpf, the number of mpo-expressing cells was significantly reduced in the MZpak4 knockdown embryos (B) compared to the uninjected control (A). Numbers refer to frequency of observed phenotype. PHENOTYPE:
|
|
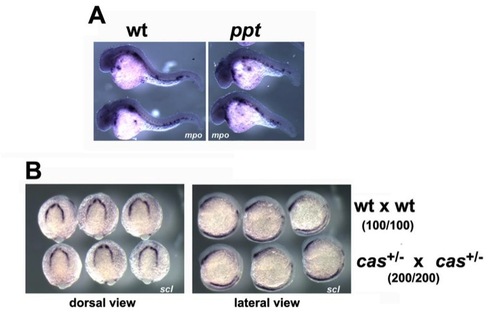
Primitive hematopoiesis is not affected by convergent extension defects or disruption of signaling from endoderm. (A) In situ hybridization with mpo in wild type (wt) and pipetail mutants (ppt, wnt5a) at 30 hpf. Lateral views shown. (B) In situ hybridization with scl in wild type (wt) and embryos from an in-cross of heterozygous casanova mutants (casta56; sox32) at 7-somite stage. Dorsal/anterior (left) and lateral (right) views are shown. Data in parentheses indicate that 100% of the cas in-cross gave normal scl expression. |
Reprinted from Mechanisms of Development, 130(2-3), Law, S.H., and Sargent, T.D., Maternal pak4 Expression is Required for Primitive Myelopoiesis in Zebrafish, 181-194, Copyright (2013) with permission from Elsevier. Full text @ Mech. Dev.