- Title
-
Incomplete splicing, cell division defects, and hematopoietic blockage in dhx8 mutant zebrafish
- Authors
- English, M.A., Lei, L., Blake, T., Wincovitch, S.M., Sood, R., Azuma, M., Hickstein, D., and Paul Liu, P.
- Source
- Full text @ Dev. Dyn.
|
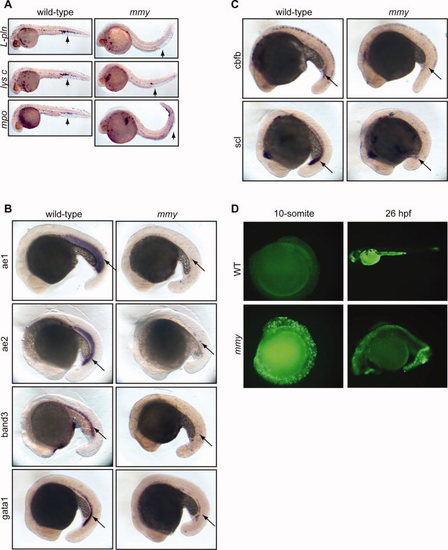
mmy mutants have severe defects in primitive hematopoiesis. A: Whole-mount in situ hybridization of myeloid markers (l-pln, lysc, and mpo) at 24 hpf. B: Expression of erythroid markers αe1 and αe2 globins, band3 and gata1 at 21 somite. C: Stem cell markers cbfb and scl are also down-regulated in mmy mutants. D: Acridine orange staining showing extensive apoptosis in mmy mutants through the entire embryos. For all probes tested, we typically used 30–40 embryos per hybridization. As such, 25% (8–10 embryos) will be mmy. EXPRESSION / LABELING:
PHENOTYPE:
|
|
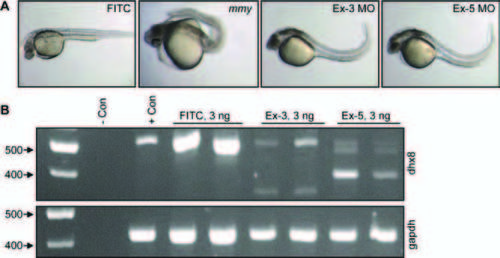
Knock-down of dhx8 by morpholino injections phenocopies mmy phenotype. A: Wild-type EK embryos were injected with 3 ng of splicing morpholinos against the donor sites in exons 3 and 5. An FITC-labeled MO was injected as a control. B: RT-PCR analysis for dhx8 from morpholino-injected embryos in A. Top panel shows that dhx8 is specifically knocked-down in embryos injected with either exon 3 or 5 MO resulting in aberrantly spliced products. The forward primer is located in exon1 and the reverse primer in exon 6. Bottom panel shows expression of gapdh as an internal control for RNA loading (Kohli et al., 2005). PHENOTYPE:
|
|
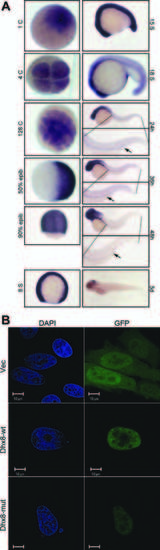
Maternal and ubiquitous expression of dhx8. A: Whole-mount in situ hybridization of dhx8 showing that dhx8 is maternally transcribed. Expression is ubiquitous to about 15–somite stage and then gradually becomes restricted to the brain as the embryo continues to develop. At 24–48 hpf, expression can be seen in the pronephric duct (arrow). B:dhx8 is expressed in the nucleus in a punctuate pattern. HeLa cells were transfected with GFP-dhx8 fusion constructs and observed by immunofluorescence 48 hr post-transfection. |
|
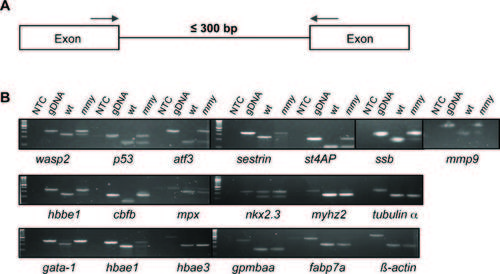
mmy mutants have a defect in mRNA splicing. A: Schematic of the PCR assay used to assess nuclear splicing. Blue arrows indicate the locations of the forward and reverse primers. B: RT-PCR analysis from 24-hpf wild-type and mmy mutant embryos was performed for the indicated genes (Supp. Table S5). Zebrafish genomic DNA (gDNA) was included to show the sizes of the un-spliced products. NTC, no template control. PHENOTYPE:
|
|
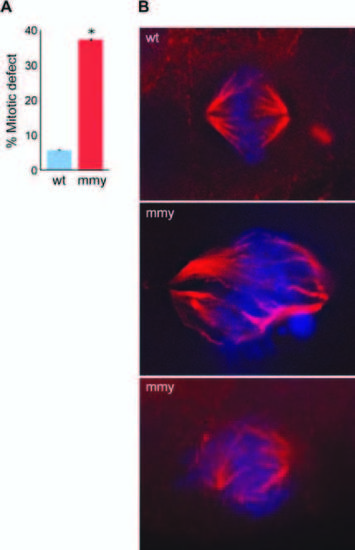
mmy mutant embryos have mitotic defects. A: Quantification of immunohistochemistry experiment. Ten wild-type and ten mutant embryos were used to count 180 mitotic events (ME) in wild type and 221 ME in mutant embryos. The embryos were sectioned into head and trunk sections and the total number of ME for each embryo counted. Error bars indicate the standard deviation between each experiment. *P = 0.000031 (Student′s t-test). B: Wild-type and mutant embryos were sectioned (10 each), mounted, and subjected to immunohistochemistry using an α-tubulin antibody (red) and counter-stain for DNA with DAPI (blue). PHENOTYPE:
|
|
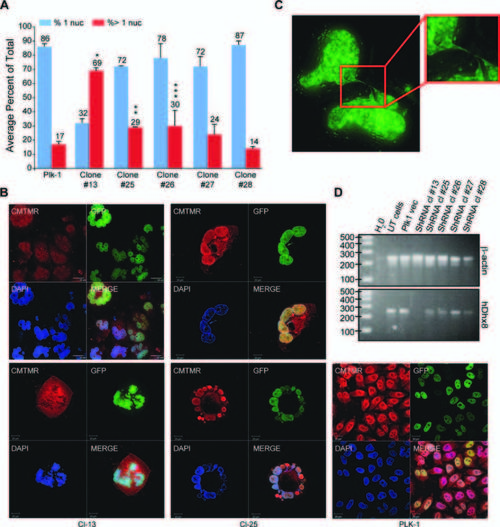
Knock-down of DHX8 in HeLa cells led to mitotic defects. A: GFP-H2B HeLa cells cultured on coverslip were transfected with 2 μg of DHX8 shRNA DNA constructs. Forty-eight hours after transfection, cells were selected in puromycin for 48 hr, fixed and subjected to confocal analysis. An average from three experiments is presented. Error bars indicate the standard deviation between each experiment. P values were calculated for the % of cells with > 1 nuclei for Plk1 control versus the different shRNA clones (Student′s t-test). *P = 0.00167, **P = 0.0149, ***: P = 0.0185. B: Transfected cells were stained with CMTMR, then subjected to confocal analysis. C: After selection, cells were subjected to Z-sectioning. One 0.2-μm slice is shown in the GFP channel. D: RT-PCR analysis using human-specific primers for ACTIN and DHX8 after the transfected cells were selected for 2 days. |
|
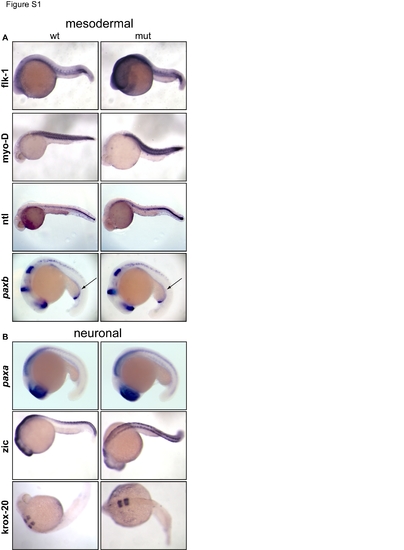
Mmy mutant embryos have no defects in mesodermal and neuronal development. (A) Whole-mount in situ hybridization of mesodermal markers at 18 somite to 24 hpf showing no difference between wt and mutant embryos. Arrows denote pronephric duct using paxb probe. (B) Whole-mount in situ hybridization of neuronal markers again showing no difference between wt and mutant embryos. |
|
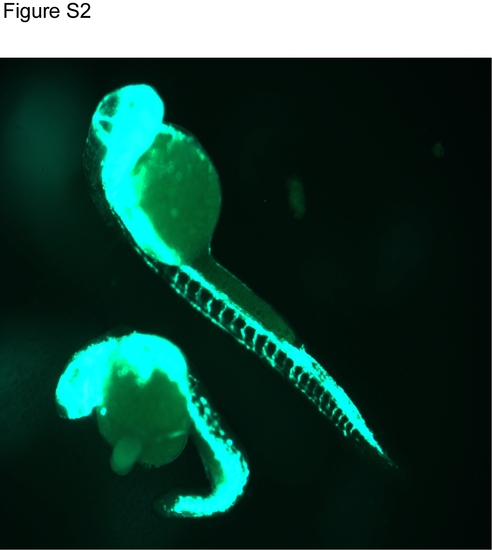
Mmy mutant embryos develop fli1-GFP+ vasculatures. Fli1-GFP embryos at 24 hpf are shown. The top embryo is a wild type embryo and the lower one is a mmy mutant. |
|
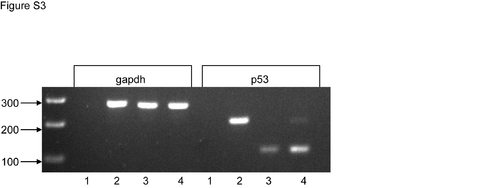
Activation of p53 in mmy mutant embryos. RT-PCR analysis from 24 hpf wild type and mmy mutant embryos showing that p53 is activated and incompletely spliced in mmy embryos. Gapdh was included as a loading control. Lane 1: H2O, lane 2: gDNA, lane 3: wt RNA, land 4: mmy RNA. |

Unillustrated author statements PHENOTYPE:
|