- Title
-
Defective cranial skeletal development, larval lethality and haploinsufficiency in Myod mutant zebrafish
- Authors
- Hinits, Y., Williams, V.C., Sweetman, D., Donn, T.M., Ma, T.P., Moens, C.B., and Hughes, S.M.
- Source
- Full text @ Dev. Biol.
|
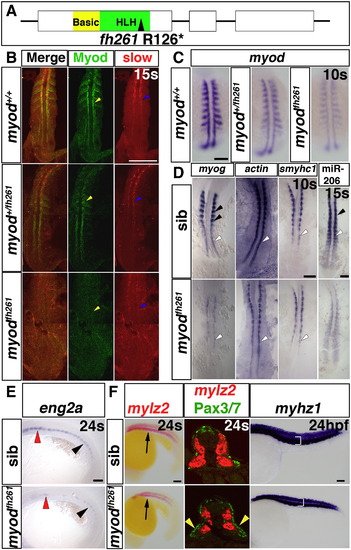
Myodfh261 mutants have delayed muscle differentiation. A. Schematic of Myod gene and protein showing the fh261 arginine to stop mutation within the helix–loop–helix domain, which is essential for dimerisation and DNA binding. B. Dual immunodetection of Myod and slow myosin in embryos from a myodfh261/+ incross, genotyped by sequencing of genomic PCR. Dorsal view, anterior to top. C. In situ mRNA hybridization reveals reduction in myod mRNA in embryos from a myodfh261/+ incross. Wholemount, anterior to top. D–F. In situ mRNA hybridization for indicated probes on myodfh261 mutants and their siblings. D. Adaxial slow precursors show reduced myog and smyhc1 at 10s and miR-206 at 15 s, but actin mRNA appeared less affected (white arrowheads). Fast muscle precursors had no myog or miR-206 expression at this stage (black arrowheads). Dorsal flatmount, anterior to top. E. eng2a expression is reduced in myodfh261 mutant in regions of muscle pioneers (black arrowhead) and medial fast fibres (red arrowhead). Lateral wholemount, anterior to left. F. Embryos labelled for fast mylz2 with Fast Red were cryo-sectioned at the region indicated (arrow, left panels). Immunofluorescent detection of Pax3/7 protein revealed increased nuclei in the lateral somite in myodfh261 mutants (yellow arrowheads). Dorsoventral extent of fast myhz1 mRNA (bracket, right panels) is reduced in myodfh261 mutants at 24 hpf. Lateral wholemounts, anterior to left. Bars = 100 μm. |
|
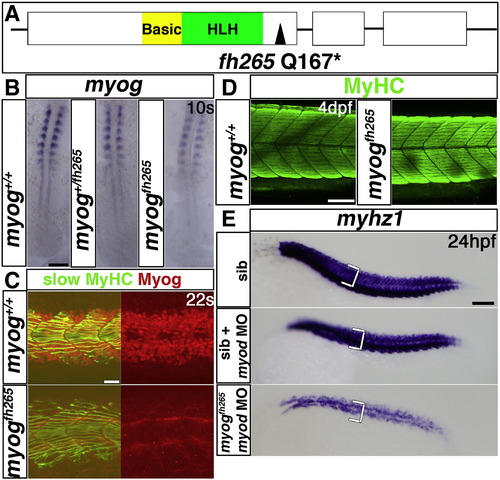
Myogenin cooperates with Myod in fast myogenesis. A. Schematic of Myog gene and protein showing the fh265 glutamine to stop mutation following the helix–loop–helix domain. B–E. In situ mRNA hybridization for myog (B) or myhz1 (E) or immunodetection of Myog and slow MyHC (C) or MyHC (D) in myog+/fh265 incrosses, injected with myod morpholino as indicated (E). B. Expression of myog mRNA is little affected in myogfh265 mutants. Dorsal flatmounts, anterior to top. C. Nuclear immunoreactivity with a polyclonal anti-rat Myog antibody is lost in myogfh265 mutants at 22 s. D. All embryos from a myog+/fh265 incross show normal levels of MyHC at 4 dpf. E. No change in myhz1 mRNA is detected in myogfh265 mutants at 24 hpf (top panel). Injection of myod MO into myogfh265 (bottom panel) leads to a greater loss of fast muscle than when injected into their siblings (middle panel), which are comparable to myodfh261 mutants (see Fig. 1F). Lateral views, anterior to left (C–E). Bars = 100 μm (except C = 20 μm). |
|
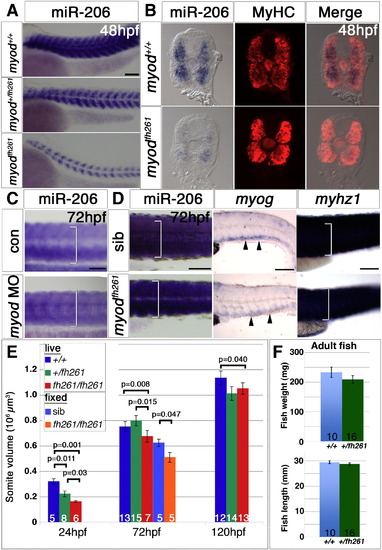
Muscle growth is impaired in myodfh261 mutants. In situ mRNA hybridization for miR-206, myog and myhz1 and immunodetection of MyHC. Lateral (A,C,D) views, anterior to left or transverse cryosections (B). A. miR-206 in sequence-genotyped 48 hpf embryos from a myod+/fh261 incross is primarily in fast fibres and is strongly reduced. B. Embryos from A were sectioned and stained with MyHC. miR-206 expression is strongly reduced in myodfh261 mutants compared with their siblings, while MyHC immunodetection is only mildly reduced. C. At 72 hpf, miR-206 has increased in myod morphants, but is still less than in siblings. D. Compared to sibs, myodfh261 mutants (identified by head muscle defects) had less myog mRNA (arrowheads) and reduction in dorsoventral extent of somitic miR-206 and myhz1 mRNA (brackets). E. Somite volume increases with age, but is reduced in myodfh261 mutant. These differences were consistent in live and fixed embryos. Number of embryos per condition is shown on columns and t-test statistics are indicated above columns. F. Adult fish mass and length. Bars = 100 μm. |
|
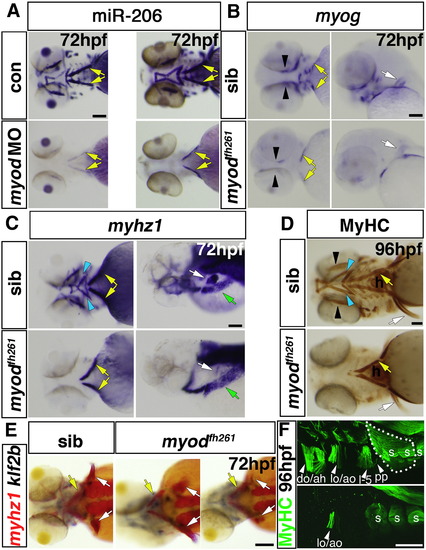
Myodfh261 mutants lack head muscles and have defects in cartilage morphogenesis. In situ RNA hybridisation for miR-206 (A), myog (B), myhz1 (C,E) and klf2b (E) or immunodetection of MyHC (MF20, D; A4.1025, F). Ventral (A,D and left panels of B and C), lateral (F, right panels of B and C) or dorsal (E) views, anterior to left. A. Myod morphant and myodfh261 mutant have similar loss of most cranial muscles except sternohyoideus (yellow arrows). B–D. In myodfh261 mutants, myog and myhz1 mRNAs and MyHC are lost from many cranial muscles (blue arrows). Expression remains in sternohyoideus (yellow arrows). Weak myog mRNA is detected in adductor mandibulae (B, black arrows) and occasional fibres differentiate in adductor mandibulae. Hypaxial muscles (C, green arrows) seem unaffected and pectoral fin muscles (B–F, white arrows, dotted area in F) are variably reduced. E. In myodfh261 mutants, most head muscles are consistently missing, even though sternohyoid (yellow arrows) and cartilage precursors expressing klf2b are present, Pectoral fin muscle is variably reduced (white arrows). F. Higher magnification shows that levator and adductor operculi are partially spared in myodfh261 mutants, whereas pectoral fin and other muscles are missing, revealing the underlying somitic muscle (s). do, dilatator operculi; ah, adductor hyomandibulae; lo, levator operculi; ao, adductor operculi; l–5, levator 5; pp, protractor pectoralis. Bars = 100 μm. |
|
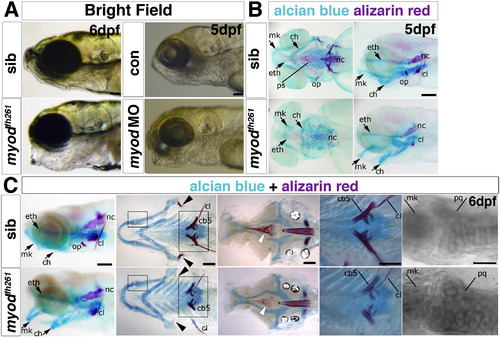
Myodfh261 mutants have cranial bone defects. A. Bright field image of a live 6 dpf myodfh261 mutant (retrospectively sequence genotyped) and a 5 dpf myod morphant showing craniofacial defects of lowered jaw and open mouth compared with sibling and control, respectively. B,C. Alcian blue and alizarin red double staining for cartilage and bone in myodfh261 mutants and siblings. At 5 dpf (B), cartilage components in mutants have different morphology with the whole lower jaw dropping, forming a gaping mouth. Both first and second arch derivatives, such as Meckel′s (mk) and ceratohyal (ch) cartilage respectively, are bent ventrally. The ethmoid plate (eth) is only mildly affected. Little bone staining is detected in the mutant. Anterior to left, ventral (left panel) and lateral (right panel) views. At 6 dpf (C), myodfh261 mutants show various cartilage and bone defects; the opercle (arrowheads) is missing, ceratobranchial 5 and the parasphenoid (white arrows) less ossified bone and the cleithrum is reduced. Cartilage cells in the mutant remain rounded, failing to mature into flattened columns. Anterior to left. Wholemounts in lateral view (left panel), or flatmounts of the dissected pharyngeal skeleton (ventral view, second from left) or neurocranium (dorsal view, central panel). Large boxed areas are magnified in the fourth panels, showing reduced ossification of ceratobranchial 5 and cleithrum. Small boxed areas are magnified in righthandmost panels to reveal rounded morphology of cartilage cells in the joint between Meckel′s and palatoquadrate (pq) cartilages in the myodfh261 mutant. cl, cleithrum; nc, notochord; op, opercle; ps, parasphenoid; cb5 ceratobranchial 5. Bars = 100 μm. |
|
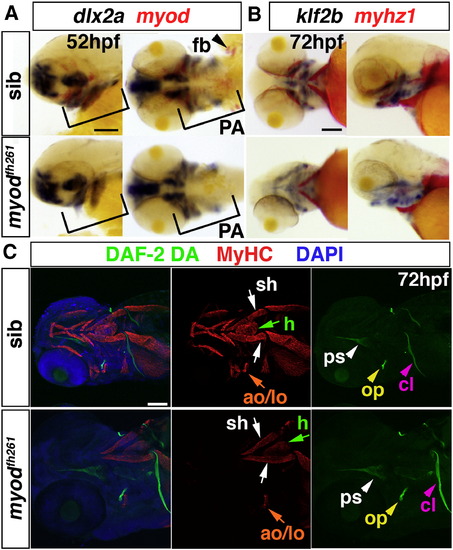
Early cranial skeletal development is normal in myodfh261 mutants. A,B. In situ mRNA hybridisation for dlx2a and myod (A), klf2b and myhz1 (B) in 52 hpf (A) and 72 hpf (B) myodfh261 mutant and sibling embryos, shown in wholemount, anterior to left. Outer panels, lateral viewand central panels, ventral view. Myod mRNA is absent but dlx2a unaffected in the pharyngeal arches (PA, brackets). Myhz1 mRNA is detected in sternohyoideus (sh, arrows) but not in other head muscles in the mutant. klf2b mRNA in skeletal parts is normal. C. Confocal stacks of heads of myodfh261 mutant and sibling stained with DAF-2DA (green), MyHC (MF20, red) and DAPI (blue). Anterior to left, ventrolateral view. Muscle in the mutant is missing except for the sternohyoideus (sh, white arrows), adductor operculi and levator operculi (ao and lo, orange arrows). Bones such as the opercle (op, yellow arrowhead), cleithrum (cl, pink arrowheads) and parasphenoid (ps, white arrowheads) are normal at this stage. Bar = 100 μm. |
|
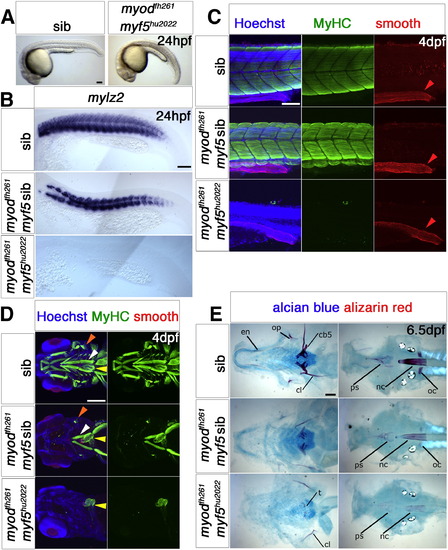
Myf5 or Myod is required for myogenesis. Wholemounts (A), flatmounts (B) and confocal stacks (C,D) in lateral (A–C) or ventral (D) view of zebrafish embryos from myf5hu2022/+;myodfh261/+ in-cross analysed at the indicated stage by in situ mRNA hybridization for mylz2 (B) or immunohistochemistry for all-MyHC (green), smooth muscle myosin heavy chain 11 (red) and Hoechst (blue) in C,D, anterior to left. A. By 24 hpf, double mutant embryos are curved and have misshapen somites with signs of cell death. B. mRNA encoding fast myosin is reduced in putativemyodfh261 mutants and absent in doubles. C. Confocal stacks of somite-16 region at 4 dpf embryos show that no skeletal muscle MyHC is detected in genotyped double mutants, in which embryos are severely reduced in size. Smooth muscle myosin in the gut (red arrowheads) are unaffected. D. Confocal stacks of the head region of 4 dpf embryos show that all skeletal muscle is missing in myf5hu2022;myodfh261 double mutant, while heart (yellow arrows) and smooth muscle are intact. myodfh261 mutant is missing all muscle except for the sternohyoideus (white arrows), adductor operculi and levator operculi (orange arrows). E. Alcian blue and alizarin red double staining for cartilage and bone in 6.5 dpf myf5hu2022;myodfh261 shows a more severe phenotype than in myodfh261 mutants and siblings. Flatmounts of the dissected pharyngeal skeleton (ventral view, left panels) or neurocranium (dorsal view, right panels). Cartilage components in double mutants stain poorly. Bone is only detected in the double mutant in cleithrum (cl) and teeth (left panel) and faintly in the notochord (nc) and parasphenoid (ps) (right panel). oc, occipitals; op, opercle; en, enteropterygoid; cb5, ceratobranchial 5. Bars = 100 μm. |
|
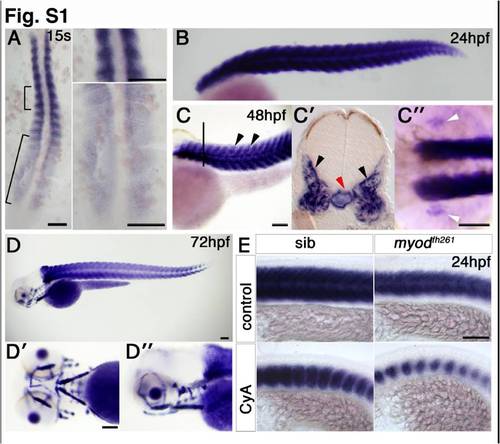
Expression of miR-206 in zebrafish development. In situ RNA hybridization for miR-206. Dorsal (A) or lateral (E) flatmounts, lateral (B, C, D, D′′), dorsal (C′′) or ventral (D′) wholemounts, anterior to top (A) or left (B–E). A. Expression is detected late in adaxial slow fibre differentiation and early in fast myogenesis. B. At 24 hpf, expression is restricted to the differentiated somitic muscle. C. By 48 hpf, expression is abundant in somitic fast muscle (arrowheads). A transverse section at the level indicated by the line in C, shows signal in somitic fast muscle (C′, black arrowheads). Notochord signal (red arrow) is detected in some embryos. Pectoral fin muscle expression is also detected (C′′, white arrowheads). D. At 72 hpf, the forming cranial muscles also contain miR-206. E. miR-206 expression is reduced in somitic muscle of myodfh261 mutant and further reduced after cyclopamine treatment, indicating that miR-206 is expressed in slow muscle fibres. Bars = 100 μm. |
|
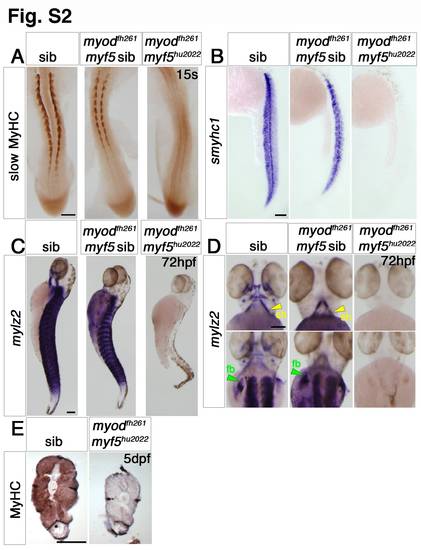
Myf5 or Myod is required for myogenesis. Dorsal flatmounts (A,D—bottom panel), ventral (D—top panel) and lateral (B,C) wholemounts or transverse cryosections (E) of zebrafish embryos from myf5hu2022/+;myodfh261/+ in-cross analysed at the indicated stage by immunohistochemistry for slow MyHC (A) or all MyHC (E) or in situ mRNA hybridization for smyhc1 (B) or mylz2 (C,D), anterior to top (A–D), dorsal to top (E). A. At 15 s, most embryos show strong slow muscle differentiation, but approximately 3/16ths (genotyped as myodfh261/fh261;myf5hu2022/+ or myodfh261/fh261;myf5+/+) have less muscle and 1/16th (genotyped as myodfh261/fh261;myf5hu2022/hu2022) have no detectable muscle. B. mRNAs encoding slow myosin is reduced in putative myodfh261 mutants and absent in doubles. C,D. At 72 hpf, no mylz2 mRNA is detected in genotyped double mutants, in which the trunk and tail are severely reduced in size. E. At 120 hpf, double mutant lacks MyHC in the gut extension region and has a reduced somite area. Note the similar sizes of non-muscle tissues. Bars = 100 μm. |
Reprinted from Developmental Biology, 358(1), Hinits, Y., Williams, V.C., Sweetman, D., Donn, T.M., Ma, T.P., Moens, C.B., and Hughes, S.M., Defective cranial skeletal development, larval lethality and haploinsufficiency in Myod mutant zebrafish, 102-12, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.