- Title
-
Essential genes for astroglial development and axon pathfinding during zebrafish embryogenesis
- Authors
- Barresi, M.J., Burton, S., Dipietrantonio, K., Amsterdam, A., Hopkins, N., and Karlstrom, R.O.
- Source
- Full text @ Dev. Dyn.
|
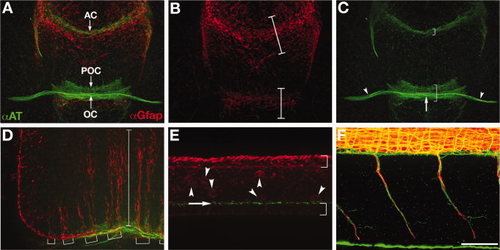
Wild-type axon and astroglial morphology. A-F: Fluorescent immunolabeling of axons (αAT, green) and astroglial cells (αGfap, red) in a 40 hpf wild-type embryo. A-C: Ventral view of the forebrain, anterior up; eyes out of view on sides. A: Combined red and green channels show the association of the forebrain commissures with glial bridges. The anterior commissure forms in the telencephalon and the post optic commissure and optic chiasm form in the diencephalon, which are separated by the optic recess. B: Gfap expressing astroglia are condensed into two “bridges”, one in the telencephalon (upper bracket) and one in the diencephalon (lower bracket; Barresi et al.,2005). C: There are three axonal pathways in the forebrain: the postoptic commissure (lower bracket), anterior commissure (upper bracket), and the optic nerves (arrowheads) that cross the midline to form the optic chiasm (arrow). D: Lateral view of the hindbrain, anterior left, dorsal up. Long radial glial processes are segmentally arranged (brackets) and span from the ventral to dorsal axis of the hindbrain (bracket). E: Lateral view of the spinal cord. This image is focused on the midline/ventricular zone of the spinal cord, with the roofplate to the top and floorplate to the bottom (brackets). Radial glial cell bodies (red, arrowheads) and cilia (green, arrow) are visible just above the floorplate. F: Lateral view of motor nerves in the trunk, anterior left, dorsal up. Axon labeling (green) is distinct from labeling of motor axon associated glia (red). αAT, anti-Aceytlated Tubulin; αGfap, anti-Glial fibrillary acidic protein; tel, (telencephalon; di, diencephalon; POC, postoptic commissure; AC, anterior commissure; OC, optic chiasm. Scale bar = 50 μm. EXPRESSION / LABELING:
|
|
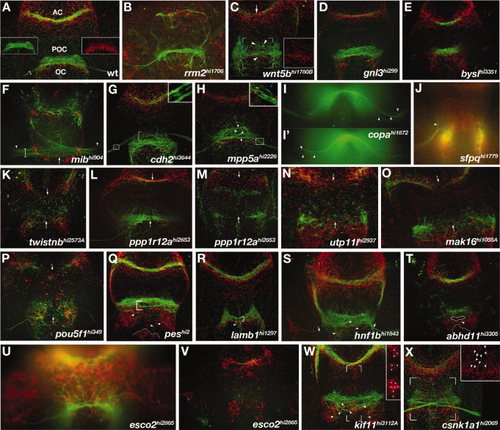
Forebrain mutants. A-X: Ventral views of zebrafish forebrains showing axons (αAT, green) and astroglial cells (αGfap, red). Anterior is up. A: WT forebrain commissures. Insets show POC (left) and the corresponding glial bridge (right) as single channels. Commissures and the optic chiasm all cross at the midline as cohesive bundles in direct association with glial bridges. B-X: Mutants with defects in forebrain astroglia and/or axonal anatomy. B: rrm2 mutants, known to have widespread necrosis, display severe astroglial reductions and dramatic optic nerve and commissural axon phenotypes including reductions and pathfinding errors. Mutants that had reduced or absent optic nerves include: (C) wnt5b, (D) gnl3, (E) bysl, (K) twistnb, (L,M) ppp1r12a, (N) utp11l, (O) mak16, (P) pou5f1,(Q) pes, (R) lamb1, (S) hnf1b, (T) abhd11, (U,V) esco2, and (W) kif11. Optic nerves were defasciculated in: (G) cdh2, (H) mpp5a, and (I) copa mutants. RGC axons made pathfinding errors in; (J) sfpq, (L) ppp1r12a, and (C) wnt5b mutants. Mutants having defasciculated commissures and/or axon pathfinding errors were; (C) wnt5b, (G) cdh2, (H) mpp5a, (L,M) ppp1r12a,(O) mak16, (P) pou5f1, (Q) pes, (R) lamb1, (S) hnf1b, (T) abhd11, (W) kif11, and (X) csnk1a1. The only mutants that did not exhibit astroglial patterning defects were (E) bysl, (G) cdh2, and (S) hnf1b. Insets show enlargements of boxed regions in the corresponding images. Arrows indicate mislocated axons phenotypes in the AC, POC (C,F,K-O), or optic nerves (S). Arrowheads highlight locations of specific axonal pathfinding errors (C,H-J,Q-T,W), astroglial defects (X), or the beginning of the optic nerves (F). White dots denote location of ectopic astoglial cell bodies at the forebrain ventricular zone (W, inset). EXPRESSION / LABELING:
PHENOTYPE:
|
|
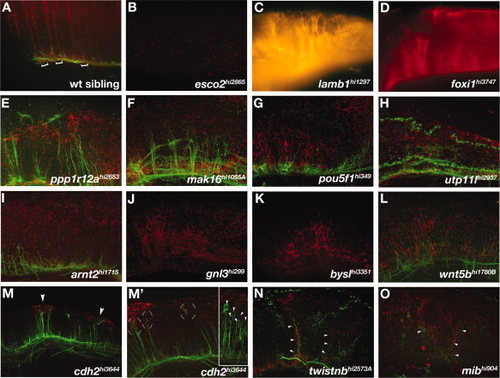
Hindbrain mutants. A-O: Lateral views of hindbrains of (A) wild-type and (B-O) mutant embryos. A: Brackets show the segmental organization of radial glial cells that extend processes dorsally to the pial surface. B:esco2 mutants had reduced radial glia in the hindbrain. C-G:lamb1, foxi, ppp1r12a, mak16, and pou5fl mutants had disorganized radial glial process that were still segmentally organized. H-L:utp11l, arnt2, gnl3, bysl, and wnt5b mutants also had disorganized radial glial processes in a generally disrupted hindbrain. M-O:cdh2, twistnb, and mib mutants had ectopic clusters of Gfap labeling. M: cdh2 mutants showed ectopic dorsal glial clusters (M, arrowheads) associated with mis-located axonal projections (M2, circular brackets). (M′) Inset shows another cdh2 mutant embryo with neuronal cell bodies associated with glial clusters (arrowheads). N,O: twistnb and mib mutants had mispositioned axons associated with mispatterned astroglia (arrowheads). B,C,D,J,K: Axon labeling is not visible due to faint fluorescent labeling. PHENOTYPE:
|
|
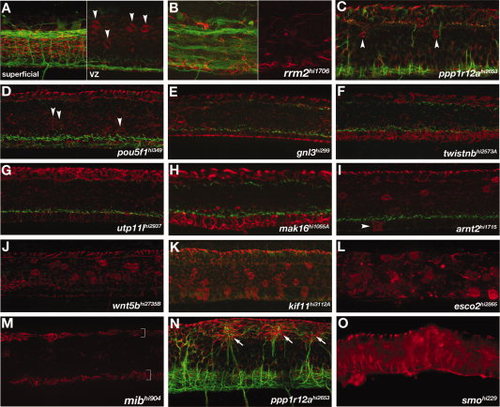
Spinal cord radial glial mutants. A-O: Lateral views of the trunk, anterior left. A: Wild-type spinal cord showing axon bundles (green) and radial glia (red) observed in a full maximum intensity projection (superficial, left panel). The typical placement and number of Gfap-labeled radial glial cell bodies at the ventricular zone (arrowheads) is seen in a projection of optical slices encompassing the full ventricular zone (“VZ”, right panel). B:rrm2 mutants had axonal and astroglial phenotypes characteristic of nonspecific defects that are likely caused by ubiquitous cell death. Right half shows only the channel for αGfap labeling (red), while the left half also includes αAT labeling for axons. C-O: Mutants with altered numbers of αGfap-labeled cell bodies within the spinal cord. C,D:ppp1r12a and pou5f1 mutants had reduced numbers of αGfap-labeled cell bodies (arrowheads) near the ventricular zone (A; see also Fig. 1E). E-H:gnl3, twistnb, utp11 and mak16 mutants had no αGfap-labeled cell bodies in the ventricular zone. I-L: In contrast, arnt2 mutants had ectopic Gfap+ cell bodies in the floorplate (I), and wnt5b, kif11, and esco2 mutants had increased numbers of Gfap+ cell bodies in the ventricular zone (J-L). M-O:mib, ppp1r12a, and smo mutants all had fewer or no αGfap-labeled cell bodies and radial glial processes. mib mutants had no radial glial processes except in the regions of the roof- and floorplates (M, brackets). N: In ppp1r12a mutants, aberrant axons projections were associated with ectopic dorsal clusters of radial processes (arrows). O: smoothened mutants had no αGfap-labeled cell bodies and generally disorganized radial glial processes. PHENOTYPE:
|
|
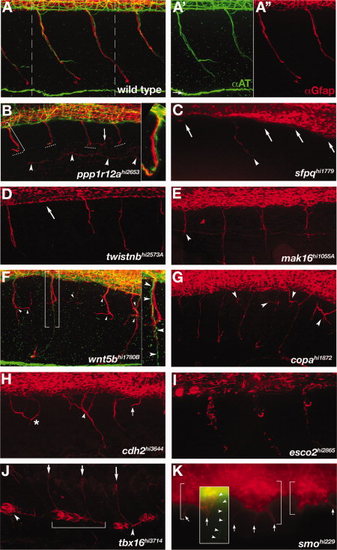
Motor nerve mutants. A-K: Lateral view of the trunk, immunolabeling shows motor axons (green) and motor axon associated glia (red). Anterior left, dorsal up. Confocal z-stacks span the region from the spinal cord exit point to the most superficial location of the spinal nerve. For technical reasons, axon labeling was only successful in ppp1r12a, wnt5bhi1780B, and smo mutants. All other mutants descriptions are based solely on αGfap labeling of motor axon associated glia. A: Wild-type spinal nerves in two full trunk segments. A2,A3: Individual axonal (green) and glial (red) labeling corresponding to the region in A between dashed lines. B: In ppp1r12a mutants motor axons and associated glia terminate (dashed lines) before reaching the lateral line (arrowheads). Motor axons (green) and glial processes (red) are tightly associated (bracket, inset). A mis-localized glial process is seen in one segment (arrow). C-E:sfpq, twistnb, and mak16 mutants had truncated (arrowheads) or absent (arrows) motor axon associated glia. F,G:wnt5b and copa mutants had excessively branched (arrowheads) motor axon associated glia. In some cases motor axons projected correctly (F, brackets, inset, arrowheads), while the associated glia were truncated and disorganized (F,G, arrowheads). H:cdh2 mutants had errors in the positioning of motor axon associated glia akin to motor axon wandering (asterisks), excessive branching (arrowhead) and variable truncations (arrow). I:esco2 mutants had reduced and fragmented motor axon associated glia. J:tbx16 mutants had ectopic glia located ventrally (arrowheads). In some somites glia spanned somite boundaries (bracket). K:smo mutants had ectopic glial clusters (brackets) with aberrant projections (arrows). An overlaid inset shows an over-intensified image of αAT labeling to make visible the rare case in which a motor axon (arrowheads) exits the spinal cord, although is seen here not associated with ectopic αGfap-labeled cell clusters (arrow). PHENOTYPE:
|
|
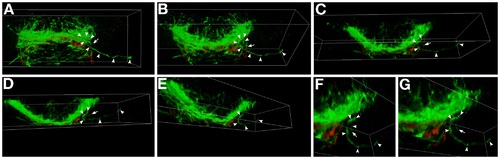
Three-dimensional analysis of optic nerve defects in ppp1r12a mutants. A-G: Selected maximum intensity projections that correspond to the 3-D rotation of the postoptic commissure and developing optic nerves. In ppp1r12a mutants at this time (40 hr) the optic nerves are seen at or near the midline, but one of the optic nerves turns away from the midline and grows ipsilateral (axon, arrowheads; leading edge, arrow). B-D: This retinal ganglion axon pathfinding error is more obvious when the forebrain is rotated downward along the X axis. E-G: Rotating along the Y axis. F,G: Higher magnification of the ipsilaterally growing axon. Axons, green (αAcetylated Tubulin); astroglia, red (anti-Gfap). This data corresponds to the movie presented in Supplemental Movie 1 |

Unillustrated author statements |