- Title
-
Neuronal Regulation of the Spatial Patterning of Neurogenesis
- Authors
- Gonzalez-Quevedo, R., Lee, Y., Poss, K.D., and Wilkinson, D.G.
- Source
- Full text @ Dev. Cell
|
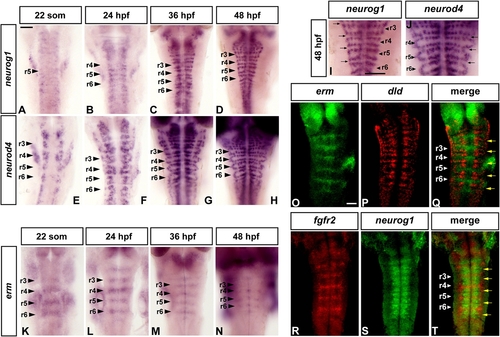
FGF Signaling Is Restricted to Nonneurogenic Regions in the Zebrafish Hindbrain |
|
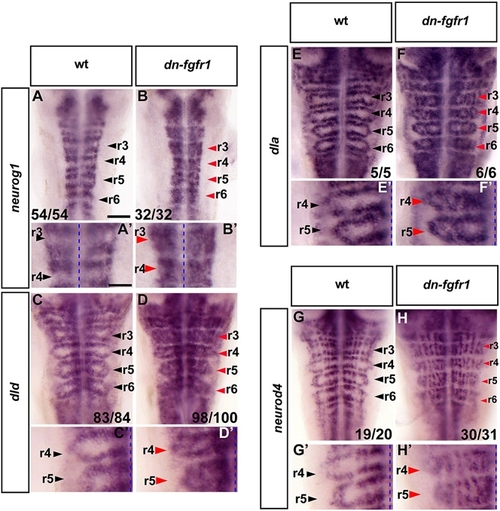
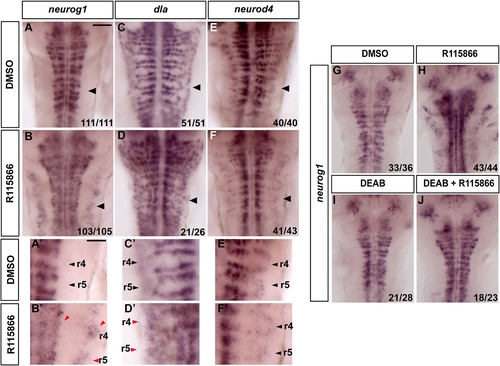
Blocking FGFR Activation Results in Proneural Gene Expression and Differentiating Neurons in Segment Centers |
|
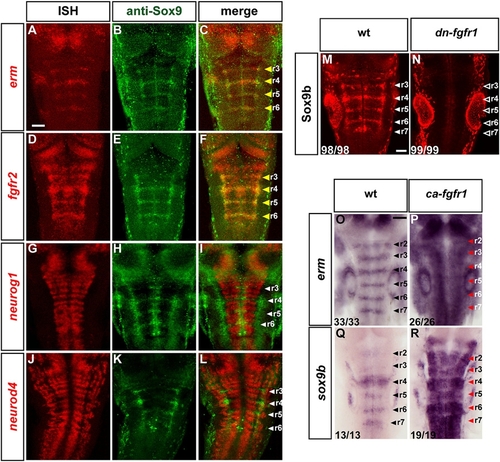
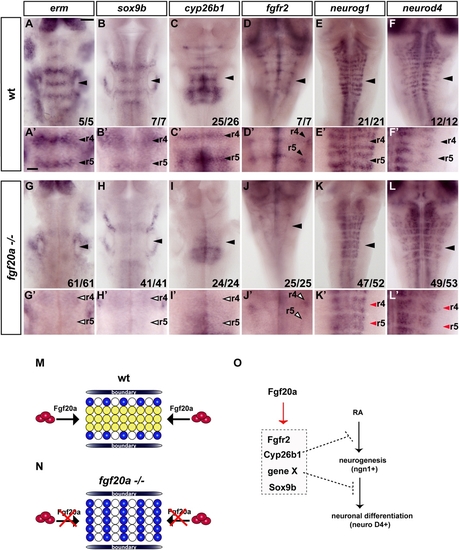
FGF Signaling Maintains a Sox9b- Expressing Population in Segment Centers |
|
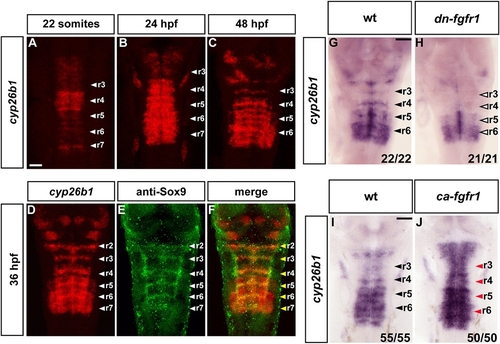
Cyp26b1 Is Expressed in Segment Centers and Regulated by FGF Signaling |
|
Blocking Cyp26 Activity Results in Premature Neurogenesis |
|
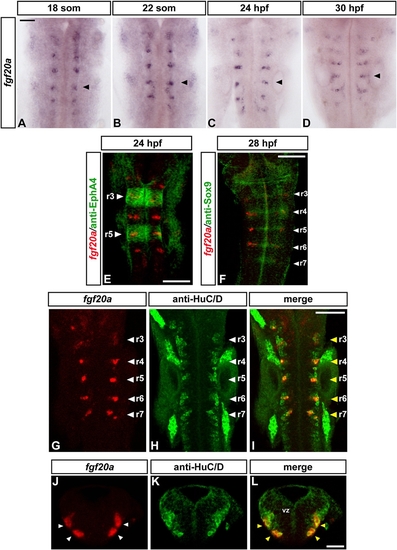
fgf20a Is Expressed in Neurons at Segment Centers |
|
fgf20a Is Required for Inhibition of Neurogenesis in Segment Centers |
Reprinted from Developmental Cell, 18(1), Gonzalez-Quevedo, R., Lee, Y., Poss, K.D., and Wilkinson, D.G., Neuronal Regulation of the Spatial Patterning of Neurogenesis, 136-147, Copyright (2010) with permission from Elsevier. Full text @ Dev. Cell