- Title
-
Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins
- Authors
- Lee, Y., Hami, D., De Val, S., Kagermeier-Schenk, B., Wills, A.A., Black, B.L., Weidinger, G., and Poss, K.D.
- Source
- Full text @ Dev. Biol.
|
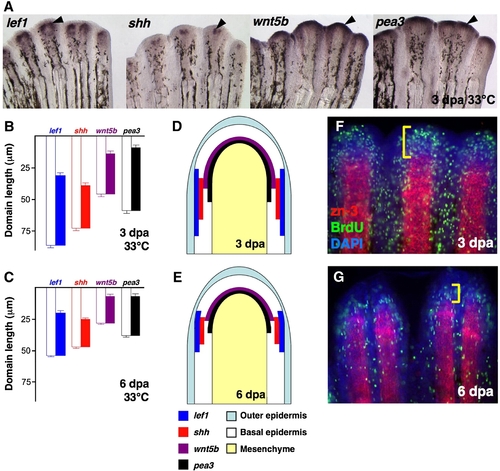
Expression of epidermal regulators in the regenerating zebrafish fin. (A) Whole-mount ISH of 3 dpa fin regenerates (33 °C) for lef1, shh, wnt5b and pea3 (Black arrowheads, violet). (B,C) Quantification of epidermal expression domains measured from either 3 dpa (B) or 6 dpa (C) whole-mount ISH-processed fins, plotted as mean ± SEM. White bars: length from distal tip of regenerate to proximal end of ISH signal; colored bars: total length of ISH expression domain. (D,E) Cartoon representation of data from (B, C), represented as a longitudinal section of 3 dpa (D) or 6 dpa (E) fin regenerates. (F,G) Whole-mount staining for BrdU (green) and scleroblasts (red) in 3 dpa (F) and 6 dpa (G) fin regenerates. Bracket indicates the proliferative blastema. |
|
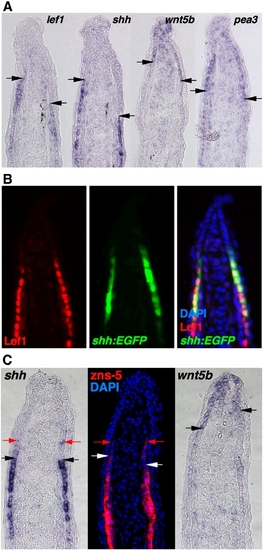
The regenerating zebrafish fin has two epidermal cellular subtypes. (A) ISH for lef1, shh, wnt5b and pea3 in serial sections from a single 3 dpa (33 °C) fin regenerate. Arrows indicate the distal end of the lef1, shh or the proximal end of the wnt5b, pea3 signals. Note that section ISH (A) gives slightly different domain representation than whole-mount ISH as in Fig. 1A, based on differences in probe penetration and signal development (Smith et al., 2008). (B) Antibody staining for Lef1 in a section from 3 dpa (33 °C) shh:EGFP fin regenerates. The distal limit of the Lef1 staining (red) is aligned with the distal limit of Shh expression (green). (C) ISH for shh, wnt5b and antibody staining for zns-5 to mark scleroblasts, using serial sections from single 3 dpa (33 °C) fin regenerate. Black and white arrows indicate the distal end of the shh and zns-5 signal, or the proximal end of the wnt5b signal. Red arrows indicate distal limits of weak signals, still similar between shh and zns-5, and non-overlapping with wnt5b. |
|
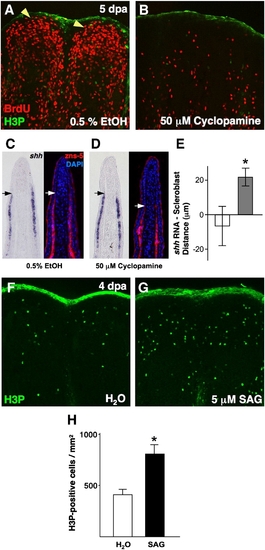
Pharmacologic manipulation of Hh signaling in ongoing regenerates affects blastemal proliferation and scleroblast patterning. (A, B) Analysis of BrdU incorporation in 5 dpa fin regenerates (26 °C) of animals treated with 50 μM cyclopamine for 28 h. Cyclopamine treatment dramatically reduced blastemal BrdU incorporation (B), as compared to vehicle-treated control animals (A; yellow arrowheads indicate blastema). (C, D) Serial sections of vehicle- (C) and cyclopamine-treated fin regenerates (D) stained for shh mRNA localization or scleroblasts (zns-5). Mesenchymal scleroblasts align with epidermal shh signals in vehicle-treated regenerates, while they significantly trail these signals after 28 h cyclopamine treatment. Arrows indicate the distal limit of detectable shh or scleroblasts. (E) Quantification of distance between distal limits of shh expression and patterned scleroblasts. White bar = vehicle; gray bar = cyclopamine (n = 11; mean ± SEM; Student′s t-test, *P < 0.05). (F, G) Images of H3P staining of 4 dpa fin regenerates (26 °C) of animals treated with 5 μM SAG or vehicle for 2 h. Activation of Hh signaling by SAG treatment enhanced the number of mitotic blastemal cells (G). (H) Quantification of mitotic counts in SAG- and vehicle-treated animals. White bar = vehicle; black bar = SAG (n = 12; mean ± SEM; Student′s t-test, *P < 0.05). |
|
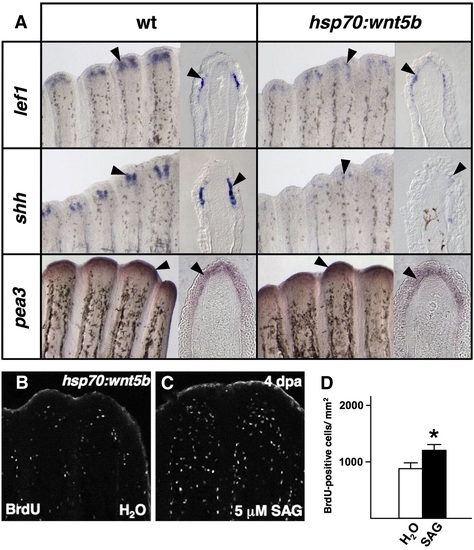
Wnt5b restricts epidermal expression of shh. (A) Whole-mount ISH and representative sections of epidermal gene expression 5 h after a single heat-shock in 4 dpa wildtype and hsp70:wnt5b fin regenerates. Ectopic overexpression of wnt5b diminishes expression of epidermal markers lef1 and shh (arrowheads), while it has no detectable effect on the Fgf target gene pea3. (B, C) Images of BrdU incorporation in 4 dpa fin regenerates of hsp70:wnt5b animals given a heat-shock, followed by SAG or vehicle treatment. SAG treatment partially rescued proliferation in hsp70:wnt5b regenerates. (D) Quantification of BrdU-positive cells in SAG- and vehicle-treated hsp70:wnt5b animals (n = 9; mean ± SEM; Student′s t-test, *P < 0.05). |
|
Transgenic Fgfr blockade depletes both positive and negative epidermal regulators. (A) Whole-mount ISH for epidermal markers, and representative sections, 5 h after a single heat-shock in 4 dpa wildtype and hsp70:dn-fgfr1 regenerates. The expression of Fgf target genes pea3 and erm is markedly reduced during Fgfr inhibition, as are other epidermal markers lef1, shh, and wnt5b (arrowheads). (B, C) Images of BrdU incorporation in 4 dpa fin regenerates of heat-shocked hsp70:dn-fgfr1 animals treated with SAG. SAG augmented blastemal proliferation in hsp70:dn-fgfr1 animals. (D) Quantification of BrdU-positive cells in SAG- and vehicle-treated hsp70:dn-fgfr1 animals (n = 12; mean ± SEM; Student′s t-test, *P < 0.005). |
|
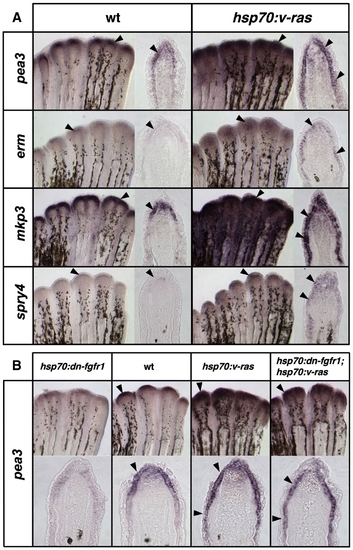
Transgenic Ras activation induces Fgf target genes in fin regenerates. (A) Whole-mount ISH and representative sections of epidermal gene expression 5 h after a single heat-shock in 4 dpa wildtype and hsp70:v-ras regenerates. Brief Ras activation increases and expands pea3 and erm expression in the basal epidermal layer, as well as the other Fgf target genes mkp3 and spry4 (arrowheads). (B) Images of whole-mount ISH and representative sections of 4 dpa regenerates from hsp70:dn-fgfr1, wildtype, hsp70:v-ras, and hsp70:dn-fgfr1; hsp70:v-ras animals, collected 5 h after a 38 °C heat-shock. While Fgfr inhibition depletes pea3 expression from the basal epidermal layer (far left), Ras activation expands pea3 expression proximally (third from left). Double-transgenic hsp70:dn-fgfr1; hsp70:v-ras regenerates (far right) display similar ectopic induction of pea3 expression as hsp70:v-ras single transgenics. |
|
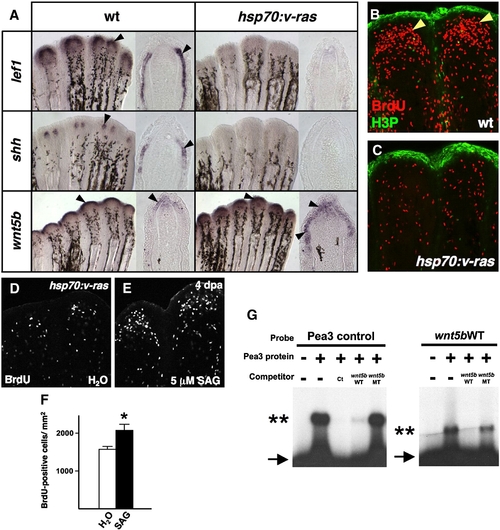
Transgenic Fgf/Ras distalizes the regeneration epidermis, blocking blastemal proliferation. (A) Whole-mount ISH and representative sections of 4 dpa regenerates from wildtype and hsp70:v-ras animals, collected 5 h after a heat-shock. lef1 and shh expression are abolished during Ras activation, while wnt5b expression is expanded. (B, C) BrdU incorporation (red) and phosphorylated Histone 3 (H3P; green) assessment in wildtype (B) and hsp70:v-ras (C) 4 dpa fin regenerates 5 h after a single heat-shock. Blastemal proliferation (arrowheads) is markedly reduced by brief Ras activation in transgenic regenerates, as compared to wildtype regenerates. (D, E) Images of BrdU incorporation in 4 dpa fin regenerates from heat-shocked hsp70:v-ras zebrafish treated with SAG or vehicle. SAG partially rescued blastemal proliferation. (F) Quantification of BrdU-positive cells in SAG- and vehicle-treated hsp70:v-ras animals (n = 12; mean ± SEM; Student′s t-test, *P < 0.05). (G) Electrophoretic mobility shift assays of binding by zebrafish Pea3 protein to a binding element upstream of the zebrafish wnt5b transcription start site. (Left) Excess addition of non-labeled wnt5b oligo (wnt5bWT) effectively competes for binding between in vitro translated Pea3 DNA-binding domain and a radiolabeled canonical binding site (Pea3 control; Ct). Arrow indicates unbound radiolabeled oligo; double asterisks marks shifted complex. A wnt5b oligo containing two nucleotide changes (wnt5bMT) fails to compete. (Right) Radiolabeled wnt5bWT mobility is shifted by the zebrafish Pea3 DNA-binding domain, an interaction competed by excess unlabeled wnt5bWT oligo. |
|
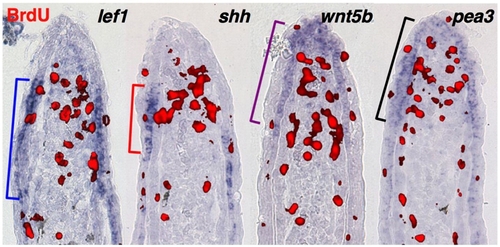
Proximal and distal epidermal territories. Representative sections from 3 dpa fin regenerates (33 °C) stained by section ISH for lef1, shh, wnt5b and pea3, and for BrdU (30 min incorporation) to mark proliferative blastemal cells (red). Brackets indicate proximal or distal limits of detectable expression. |
|
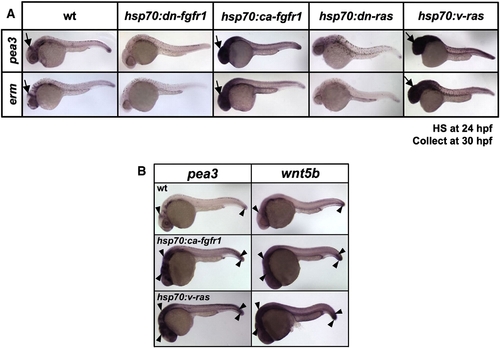
Transgenic activation of both Fgf and Ras induces Fgf target genes and wnt5b in embryos. (A) Whole-mount ISH of 30 hpf embryos from hsp70:dn-fgfr1, hsp70:ca-fgfr1, hsp70:dn-ras, or hsp70:v-ras transgenic lines that had been heat-shocked at 24 hpf. The expression of Fgf target genes pea3 and erm decreases in transgenic embryos with Fgfr or Ras inhibition, while their expression is increased and expanded in transgenic embryos in which Fgfr or Ras activity is boosted. Arrows indicate midbrain–hindbrain boundary. (B) Whole-mount ISH of 30 hpf embryos from wildtype and hsp70:ca-fgfr1 or hsp70:v-ras transgenic lines, that had been heat-shocked at 24 hpf. pea3 and wnt5b have similar expression domains in 30 hpf wildtype embryos (arrowheads), and expression of each is increased and expanded upon ectopic activation of Fgfr1 or Ras in transgenic embryos. |
|
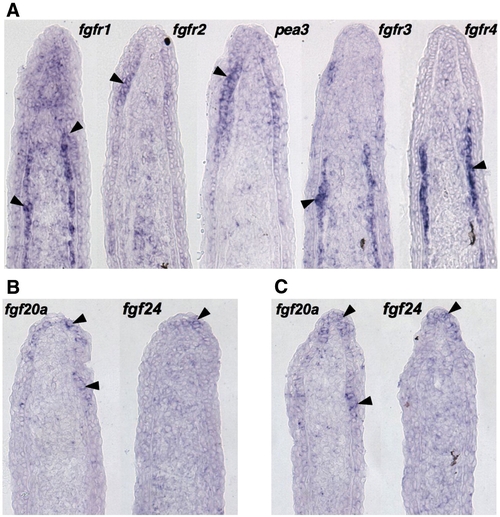
Expression patterns of fgfr and fgf genes during fin regeneration. (A) ISH of serial sections for fgfr1–4 and pea3 from a single 3 dpa (33 °C) fin regenerate, showing differential expression of receptors. fgfr1 is expressed broadly in the basal epidermis, mesenchyme, and scleroblasts, fgfr2 in the basal epidermis, and fgfr3 and fgfr4 predominantly in scleroblasts. (B, C) Two representative serial sections from 3 dpa fin regenerates (33 °C) stained by section ISH for fgf20a and fgf24. While each ligand is expressed in the basal epidermal layer, fgf20a expression extends more proximally than fgf24. |
Reprinted from Developmental Biology, 331(2), Lee, Y., Hami, D., De Val, S., Kagermeier-Schenk, B., Wills, A.A., Black, B.L., Weidinger, G., and Poss, K.D., Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins, 270-280, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.