- Title
-
Nodal signalling imposes left-right asymmetry upon neurogenesis in the habenular nuclei
- Authors
- Roussigné, M., Bianco, I.H., Wilson, S.W., and Blader, P.
- Source
- Full text @ Development
|
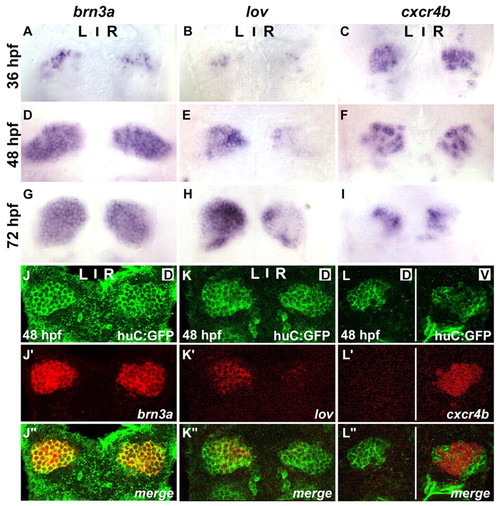
cxcr4b is expressed in the habenulae. (A-I) Expression of brn3a (A,D,G), lov (B,E,H) and cxcr4b (C,F,I) in habenulae of wild-type zebrafish embryos at 36 (A-C), 48 (D-F) and 72 (G-I) hpf. (J-L″) Single confocal images of 48 hpf Tg(huC:gfp) embryos showing immunolabelled GFP protein (J-L) and the localisation of mRNA encoding brn3a (J′), lov (K′) or cxcr4b (L′). (J″-L″) Merged images. Dorsal views of embryos with anterior at the top. Single confocal sections correspond to dorsal (D) or ventral (V) z-sections through the habenular nuclei. EXPRESSION / LABELING:
|
|
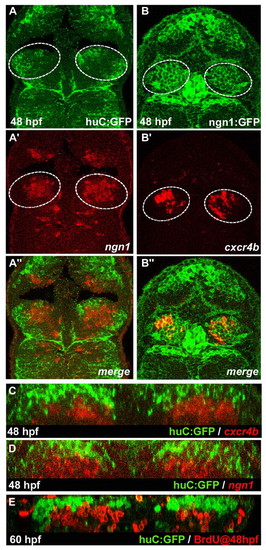
cxcr4b is a marker of habenular progenitors/early-born neurons. (A-B″) Single confocal images (dorsal views) of a 48 hpf Tg(huC:gfp) zebrafish embryo (A-A″) with in situ labelling of ngn1 and a 48 hpf Tg(ngn1:gfp) embryo (B-B″) with in situ labelling of cxcr4b. (A″-B″) Merged images. The habenulae are indicated by a dashed white line. (C-E) Cross-sections of double-labelled habenulae generated from confocal z-series. (C) Immuno-detection of GFP protein (green) and localisation of cxcr4b transcripts (red) in a 48 hpf Tg(huC:gfp) embryo. (D) Immuno-detection of GFP protein (green) and localisation of ngn1 mRNA (red) in a 48 hpf Tg(huC:gfp) transgenic embryo. (E) Immuno-detection of BrdU (red) and GFP protein (green) in a 60 hpf Tg(huC:gfp) embryo pulsed with BrdU at 48 hpf. |
|
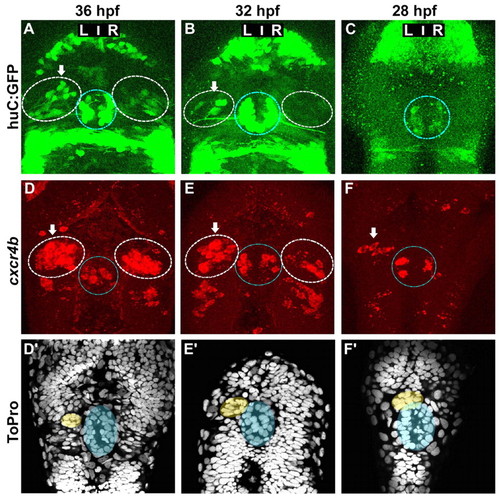
Asymmetric cxcr4b expression precedes the migration of the parapineal and the appearance of HuC+ habenular neurons. (A-C) Confocal projections of GFP fluorescence in Tg(huC:gfp) zebrafish embryos at 36 (A), 32 (B) and 28 (C) hpf. (D-F′) Confocal projections of fluorescent in situ hybridisations showing cxcr4b transcripts (D-F) and ToPro nuclear staining in the corresponding embryos (D′-F′) at 36 (D,D′), 32 (E,E′) or 28 (F,F′) hpf. Nuclear staining permits detection of the parapineal (yellow) and pineal (blue). In A-F, the habenulae are indicated by a dashed white line, the epiphysis by a blue line. Arrows indicate the habenula with the higher number of huC:GFP-positive habenular neurons or cxcr4b-expressing cells. |
|
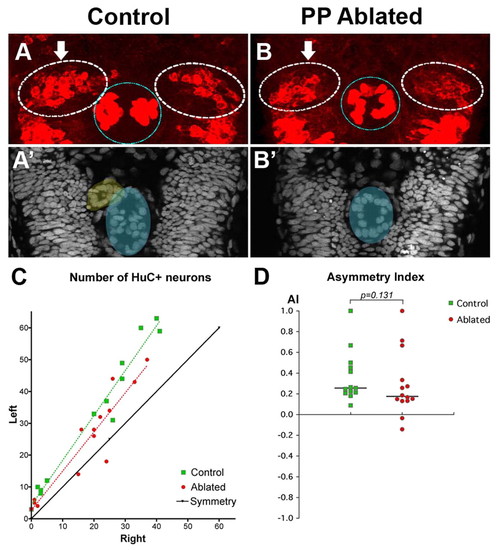
Asymmetric habenular neurogenesis is independent of the parapineal. (A,B) Confocal projection of immuno-detected HuC/D proteins in 36-38 hpf control (A) and parapineal-ablated (B) zebrafish embryos. Arrows indicate the habenula with the higher number of HuC/D+ neurons. The habenulae are indicated by a dashed white line, the epiphysis by a blue line. (A′,B′) ToPro nuclear staining of the embryos in A,B reveals the parapineal in the control (yellow) but not in the parapineal-ablated embryo; the epiphysis is shaded blue. (C) Numbers of left versus right HuC+ habenular neurons with each point representing one embryo. A symmetry line (black) is included as a reference. The dotted lines represent the best-fit linear regression for controls and ablated embryos. (D) Asymmetry index (AI, see Materials and methods) plotted for each control or parapineal-ablated embryo. The horizontal black lines indicate the median AI value for the group of like-treated embryos. |
|
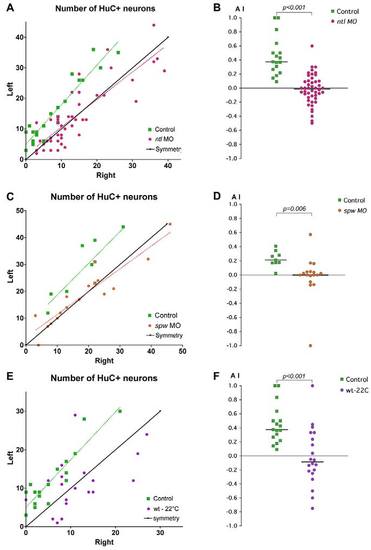
Lateralised habenular neurogenesis is compromised by treatments that disrupt Nodal signalling. Plots of the numbers of HuC+ neurons in the left and right habenulae and corresponding asymmetry indices (AI) in ntl morphants (A,B), spaw morphants (C,D) and cold-treated zebrafish embryos (E,F). (A,C,E) Habenular neurons were detected in Tg(huC:gfp) transgenic embryos using either anti-GFP (A,E) or HuC/D (C) antibody. A symmetry line (black) is included as a reference. Dotted lines represent the best-fit linear regression for each experimental condition. (B,D,F) AI values plotted for all embryos. The same control dataset is plotted for the ntl MO and cold-treatment experiments. Applying the Bonferroni correction for multiple comparisons shows that both the cold-treated and ntl MO datasets are significantly different from the control at the 5% confidence level. Horizontal black lines indicate the median AI value for the group of like-treated embryos. |
|
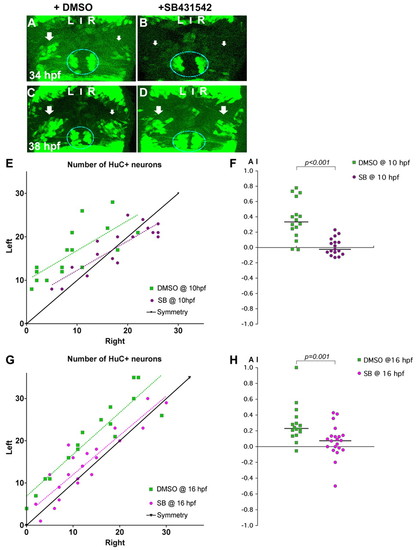
Nodal signalling promotes early asymmetric neurogenesis. (A-D) Confocal projections of Tg(huC:gfp) transgenic zebrafish embryos incubated with DMSO (A,C) or with SB431542 (B,D) from 10 hpf and fixed at 34 (A,B) or 38 (C,D) hpf. Arrows indicate that the left pools of HuC+ neurons are bigger than those on the right in DMSO-treated embryos (A,C), whereas the left and right pools are symmetric in embryos incubated with SB431542 (B,D). The epiphysis is indicated by a blue line. Habenular neurogenesis is also delayed in SB431542-treated embryos. (E-H) Numbers of left and right HuC+ habenular neurons (E,G) and corresponding AI (F,H) in embryos treated with DMSO or SB431542 at 10 (E,F) or 16 (G,H) hpf. Habenular neurons were detected in Tg(huC:gfp) transgenic embryos using an anti-GFP (E) or HuC/D (G) antibody. A symmetry line (black) is included as a reference in E and G. Dotted lines represent best-fit linear regressions. Horizontal black lines in F and H indicate the median AI value for the group of like-treated embryos. |
|
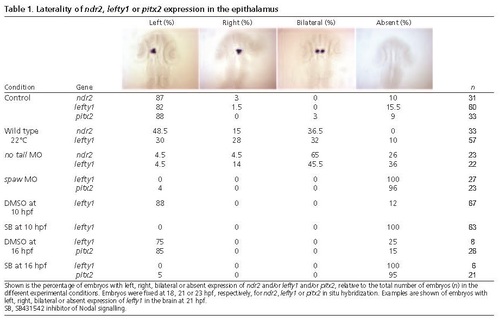
Laterality of ndr2, lefty1 or pitx2 expression in the epithalamus. Shown is the percentage of embryos with left, right, bilateral or absent expression of ndr2 and/or lefty1 and/or pitx2, relative to the total number of embryos (n) in the different experimental conditions. Embryos were fixed at 18, 21 or 23 hpf, respectively, for ndr2, lefty1 or pitx2 in situ hybridization. Examples are shown of embryos with left, right, bilateral or absent expression of lefty1 in the brain at 21 hpf. SB, SB431542 inhibitor of Nodal signalling. PHENOTYPE:
|