- Title
-
Formation of posterior cranial placode derivatives requires the Iroquois transcription factor irx4a
- Authors
- Feijoo, C.G., Saldias, M.P., De la Paz, J.F., Gómez-Skarmeta, J.L., and Allende, M.L.
- Source
- Full text @ Mol. Cell Neurosci.
|
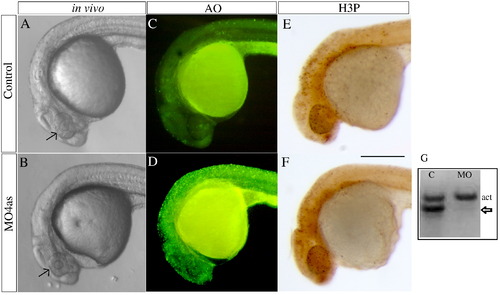
irx4a morphant embryos show defects in head size and extensive cell death. (A, B) Images of live wild type (A) and morphant (B) embryos at 24 hpf; Compare the size of the eyes and head (arrows). (C, D) Acridine orange staining of wild type (C) and irx4a morphant (D) embryos viewed at 24 hpf. Note the increased number of labeled cells in the morphant embryo (E, F) MO4as injected embryos do not show substantial changes in cell proliferation as evidenced by anti-phospho-histone H3 (H3-P) immunostaining. (G) RT-PCR showing the effect of the MO4as on irx4a transcript splicing. RNA prepared from uninjected embryos (“C”) shows the internal control band (actin, act) but the absence of unprocessed irx4a mRNA. The presence of this band in MO4as injected fish (arrow) indicates defective irx4a mRNA splicing. Panels A–F show lateral views of 24 hpf embryos; dorsal is up, anterior is left. Scale bar in F: 70 μm. |
|
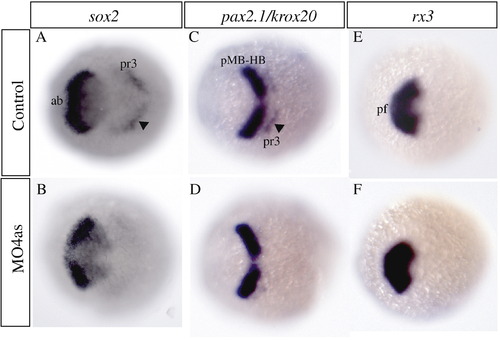
irx4a regulates sox2 expression in the neural plate. In situ hybridization of brain markers was carried out on control and irx4a morphant embryos at 90% epiboly. (A, B) The neural precursor marker sox2 appears diminished in the anterior brain (ab) and hindbrain (presumptive rhombomere 3, arrowhead, pr3) in irx4a morphant embryos (B) compared to uninjected controls (A). (C, D) The expression pattern of pax2.1 in the midbrain–hindbrain boundary (pMB-HB) appears unaffected but the expression of the rhombomere 3 marker krox20 (arrowhead) is diminished in morphant embryos. (E, F) rx3 expression in the presumptive forebrain (pf) is unchanged in morphant compared to control embryos. All views are dorsal, anterior is towards the left. EXPRESSION / LABELING:
|
|
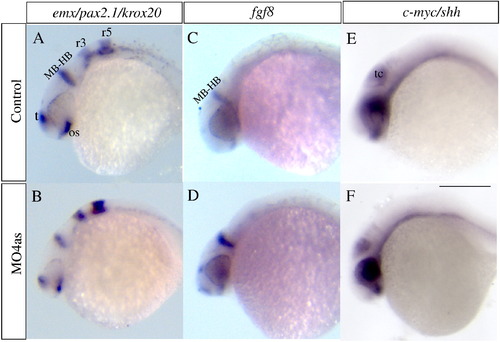
irx4a inhibition does not significantly alter A/P or D/V patterning. In situ hybridization was carried out on 24 hpf embryos using CNS markers to evaluate potential patterning defects induced by inhibition of irx4a function. (A, B) At 24 hpf the expression patterns of the forebrain marker emx, the midbrain–hindbrain boundary marker pax2.1 and the hindbrain marker krox20 appear unaffected in morphant embryos compared to controls. (C, D) Expression of fgf8 corroborates that the midbrain–hindbrain boundary is correctly established in morphant embryos. (E, F) Expression of shh and c-myc is unchanged in morphant embryos indicating that dorso-ventral patterning is also normally established in the absence of irx4a function. MB-HB, midbrain–hindbrain boundary; os, optic stalk; r3, rhombomere 3; r5, rhombomere 5; t, telencephalon; tc, tectum. The panel shows lateral views; dorsal is up, anterior is towards the left. Scale bar in (F) 50 μm. EXPRESSION / LABELING:
|
|
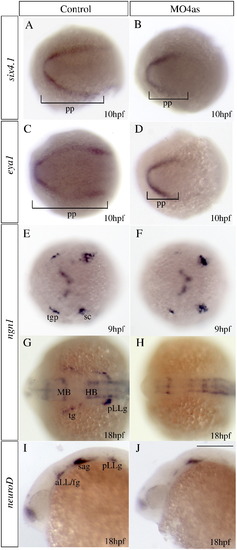
irx4a is involved in specification of a subset of placodes. In situ hybridization results of embryos processed for detecting expression of the indicated marker genes. Embryos were fixed at 10 hpf (A–F), 17 hpf (G–H) or 24 hpf (I–J). (A–D) Injection of MO4as leads to an inhibition of the posterior expression domain of the preplacodal markers six4.1 (compare A and B) and eya1 (compare C and D). (E–H) ngn1 expression is strongly diminished at 9 hpf in the trigeminal ganglion (tg) but not in the posterior lateral line ganglion (pLLg) in morphants (compare E to F). Later in development, at 17 hpf, expression of this marker is absent in the tg and pLLg (compare G to H). Expression of the proneural gene neuroD in the anterior and posterior lateral line ganglia is also affected in morphants (compare I to J). pp, preplacodal region; aLL, anterior lateral line placode; HB, hindbrain; MB, midbrain; sc, dorsal/lateral spinal cord; sa, statoacoustic ganglion; pLL, posterior lateral line placode; pLLg, posterior lateral line ganglion; tgp, trigeminal ganglion placode; tg, trigeminal ganglion. A–H are dorsal views; I, J show lateral views; anterior is towards the left. Scale bar in J: 40 μm for G–J. |
|
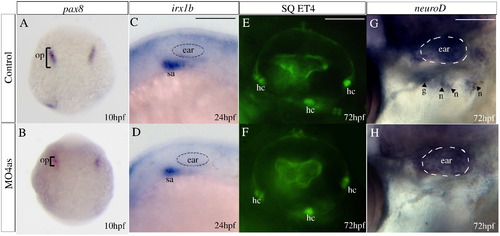
Requirement of irx4a for epibranchial ganglia but not ear development. In situ hybridization results (A–D, G–H) and images of GFP expression in transgenic larvae (E–F) for control and irx4a morphant fish. (A, B) Expression of the otic placode marker pax8 is slightly decreased in irx4a morphant embryos at 10 hpf. (C, D) Expression of the statoacoustic ganglion marker irx1b is similarly affected at 24 hpf. (E, F) Examination of hair cells (hc) in control and morphant SqET4 transgenic larvae shows no effect of the morpholino injection at 72 hpf. In this transgenic line, the three patches of hair cells in the ear can be observed in live fish. (G–H) In situ hybridization showing neuroD expression in the epibranchial ganglia (petrosal, p, and nodose, n) at 72 hpf. Note the absence of expression of this marker in morphant (H) compared to controls (G). hc, clusters of hair cells; n, nodose ganglia; sa, statoacoustic ganglion; o, otic placode; p, petrosal ganglion. A, B dorsal views of whole embryos. C–H lateral views centered on the otic vesicle. Scale bar in C: 15 μm for C, D; scale bar in E: 5 μm for E, F; scale bar in I: 10 μm for G, H. EXPRESSION / LABELING:
|
|
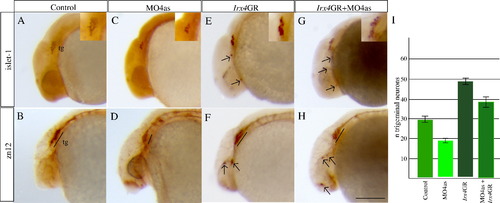
irx4a regulates the number of neurons that form the trigeminal ganglion. irx4a morphant and control embryos were fixed at 24 hpf and processed for immunohistochemistry using the anti-Islet1 (A, C, E, G, insets show close-up images of the trigeminal area) or zn-12 antibodies (B, D, F, H). (A–D) Comparison of both antibody stains between control (A, B) and irx4a morphant (C, D) shows a drastic decrease in the number of neurons that form the trigeminal ganglion (compare A with C and B with D). (E, F) Gain of function experiments carried out by injection of Irx4GR mRNA show an increase in the number of trigeminal neurons (compare A with E and B with F) and occasional appearance of ectopic expression of both antibody markers in other areas of the head (arrows). (G, H) Coinjection of MO4as and Irx4GR mRNA recovers near-normal numbers of neurons in the trigeminal ganglion though there is remaining ectopic expression of Islet-1 and zn-12 antigen in the head. I, the number of anti-Islet1 labeled neurons in the trigeminal ganglia were counted in control, MO4as, Irx4GR and MO4as+Irx4GR embryos. An average for 30 embryos with S.D. was calculated. A–H show lateral views of the anterior region of embryos; dorsal is up, anterior is left. Scale bar in H: 40 μm. |
|
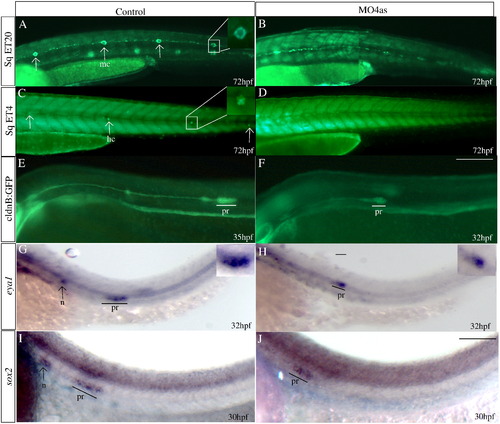
irx4a is required for the correct development of the posterior lateral line. GFP expression was observed in the SqET20 (A, B), SqET4 (C, D) and cldnB:GFP (E, F) transgenic lines in control and morphant larvae at the indicated times post-fertilization. In situ hybridization was carried out using the eya1 (G, H) and sox2 (I, J) riboprobes. (A, B) Expression of GFP in 72 hpf control SqET20 transgenic line labels mantle cells (arrows in A, inset) and interneuromastic cells (inc). Morphant embryos (B) lack neuromast mantle cells but interneuromastic cells are present indicating that primordium migration occurred. (C, D) At 72 hpf mechanosensory hair cells (arrows in C, inset) are labeled in the SqET4 transgenic line while morpholino injected siblings (D) show the absence of these cells. (E, F) The migrating primordium, the trail of interneuromastic cells and deposited neuromasts express GFP in cldnB:GFP transgenic uninjected embryos (E). In contrast, MO4as injected cldnB:GFP embryos (F) show a decrease in the size of the migrating primordium and the absence of deposited neuromasts. Note that we have compared GFP label in a 32 hpf morphant with a 35 hpf control embryo. The primordium size difference is evident despite the fact that the primordium in the control fish has already deposited two neuromasts. (G–J) Expression of the primordium markers eya1 (G, H, insets show magnified views) and sox2 (I, J) shows a similar effect, the label is diminished in morphant embryos. inc, interneuromastic cells hc, hair cells; n, neuromast; pr, primordium. The entire panel shows lateral views of the trunk and tail region; dorsal is towards top, anterior is towards left. Scale bar in F: 50 μm for A–F, scale bar in J: 30 μm for G–J. EXPRESSION / LABELING:
|
|
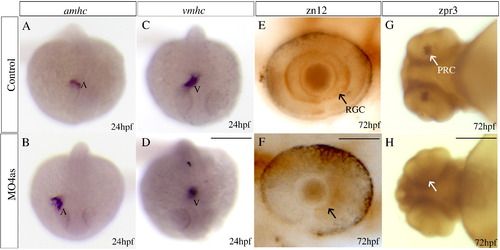
irx4a is involved in cardiac chamber specification and retinal development. Control and irx4a morphant embryos were processed for in situ hybridization (A–D) or immunohistochemistry (E–H). (A–D) Morphant embryos (B, D) show an increase in the size of the atrium and a decrease in the size of the ventricle compared to control embryos (A, C) as revealed the expression of the amhc (A, B) and vmhc (C, D) markers, respectively. (E, F) zn12 immunostaining labels retinal ganglion cells (RGC) in control embryos (E, arrow) but they are not detected in MO4as injected embryos (F). (G, H) zpr-3 immunostaining labels photoreceptor cells (PRC) in the 72 hpf control larva (G, arrow) but not in the morphant larva (H); note that photoreceptors have only differentiated in the ventral retina at this stage. A–D, dorsal views of whole embryos at 24 hpf, anterior is down; E, F, lateral views of the eye at 72 hpf, anterior is left; G, H, ventral views of the head. Scale bar in D: 40 μm for A–D, scale bar in F: 7 μm for E, F; scale bar in H: 15 μm for G, H. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Expression of irx4a at early development. Embryos were fixed and processed for in situ hybridization with an irx4a specific probe. A, At 70% epiboly (mid-gastrulation) irx4a expression is detected in the entire embryo, shield is to right and animal pole is up. |
|
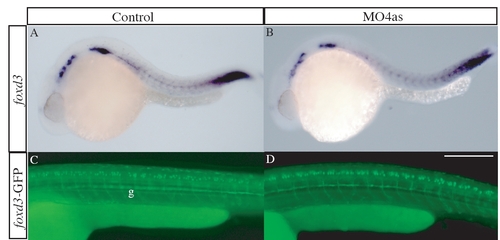
irx4a has no role in neural crest development. The expression pattern of the neural crest marker foxd3 was analyzed by in situ hybridization (A, B) and GFP expression in the transgenic line foxd3-GFP (C, D) was analyzed in control (A, C) and morphant embryos (B, D) at 24 hpf and 48 hpf, respectively. We detected no differences in the distribution or intensity of both markers. g, glia. All panels show lateral views; dorsal is up, anterior is left. Scale bar in D: 30 μm for C, D. |

Unillustrated author statements |
Reprinted from Molecular and cellular neurosciences, 40(3), Feijoo, C.G., Saldias, M.P., De la Paz, J.F., Gómez-Skarmeta, J.L., and Allende, M.L., Formation of posterior cranial placode derivatives requires the Iroquois transcription factor irx4a, 328-337, Copyright (2009) with permission from Elsevier. Full text @ Mol. Cell Neurosci.