- Title
-
Specification of vertebrate slow-twitch muscle fiber fate by the transcriptional regulator Blimp1
- Authors
- Liew, H.P., Choksi, S.P., Wong, K.N., and Roy, S.
- Source
- Full text @ Dev. Biol.
|
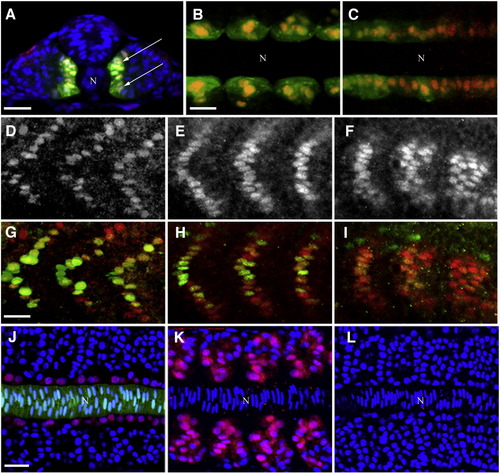
Expression of Blimp1 protein precedes the onset of slow muscle differentiation. (A–C) Blimp1 (red) is expressed and localized in the nuclei of adaxial cells (arrows) of 14 hpf zebrafish embryos labeled with GFP from the actin::gfp transgene. Blimp1 is expressed in the adaxial cell of the somites (B) as well as the presomitic mesoderm (C). (D–F) Blimp1 protein expression recedes in the more anterior somites (D, E) and is stronger in the newly formed posterior somites (F) in 18 hpf wild-type embryos. (G–I) Double staining of Blimp1 (red) and Prox1 (green) reveals reduced levels of Blimp1 in anterior somites (G, H) where Prox1 is highly expressed, while Prox1 is barely detectable in slow muscle cells of posterior somites (I), where Blimp1 is strongest. (J) Adaxial expression of Blimp1 (red) in a wild-type 14 hpf embryo immediately adjacent to the notochord (green, highlighted by GFP expression from the shh::gfp transgene). (K) Ectopic expression of Blimp1 is induced throughout the somites by constitutive activation of the Hh signaling pathway in response to dominant negative Protein Kinase A mis-expression in a 14 hpf wild-type embryo. (L) Blimp1 expression is absent in a 14 hpf smo mutant embryo deficient in Hh signaling. Nuclei in panels A, J–L were stained with DAPI (blue). In panels A–C and J–L, “N” marks the position of the notochord. Panel A depicts transverse section; panels B, C and J–L depict flat mounted embryos; panels D–I show laterally mounted embryos. In this and subsequent figures, all laterally and flat mounted embryos are oriented anterior to the left; dorsal is on top for embryos depicted in transverse sections. Scale bars = 25 μm. The scale bar in B applies to C; the scale bar in G applies to D–I and the scale bar in J applies to K and L. EXPRESSION / LABELING:
|
|
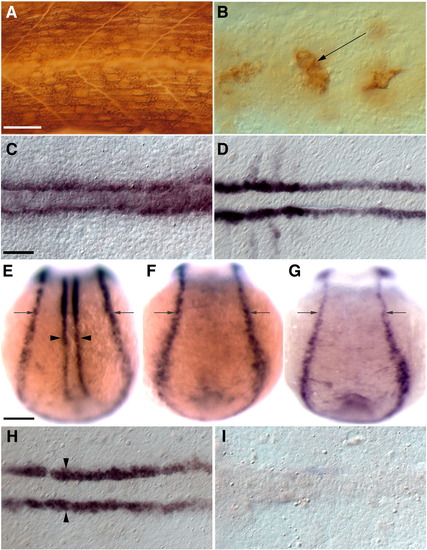
Induction of blimp1 expression by Hh signaling in presumptive slow muscle precursors requires their prior commitment to the myogenic fate. (A) MAb A4.1025 staining for all MyHC in a 24 hpf wild-type embryo. (B) MAb A4.1025 staining of a myoD; myf5 double morphant embryo showing few differentiated muscles within the somites at 24 hpf (arrow, n = 20/20). (C, D) myoD; myf5 double morphant embryos retain functional Hh signaling as demonstrated by normal expression of the Hh target genes ptc1 (C, n = 16/16) and myoD (D, n = 16/16) in adaxial cells. (E, H) Wild-type embryos showing blimp1 expression in the adaxial cells (arrowheads) and the neuro-ectodermal border (arrows). (F) blimp1 expression is absent from the adaxial cells of a smo mutant embryo. (G, I) blimp1 expression is specifically lost from adaxial cells in myoD; myf5 morphants while it appears unaffected in the neuro-ectodermal border (arrows, n = 23/23). Panels A and B depict lateral views; C–I depict dorsal views of 11 hpf embryos. C, D, H, I depict flat mounts; E–G depict whole mounts. Scale bars = 25 μm in A and C, and 100 μm in E. The scale bar in A applies to B; the scale bar in C applies to D, H and I and the scale bar in E applies to F and G. EXPRESSION / LABELING:
|
|
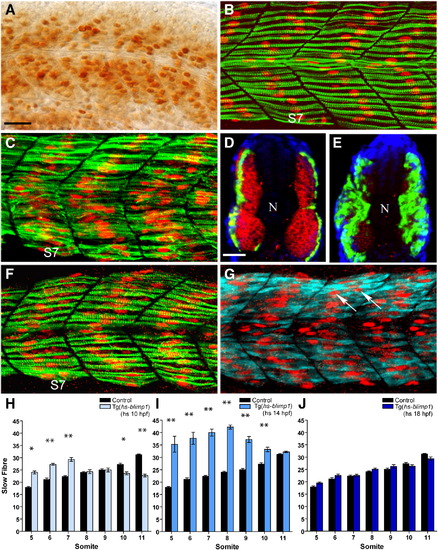
Competence of somitic myoblasts to respond to Blimp1 and adopt the slow fate changes as a function of time. (A) High level of Blimp1 protein expression (arrows) throughout the myotome in a heat induced Tg(hs::blimp1) transgenic embryo. (B) Slow muscle fibers in an uninduced 24 hpf Tg(hs::blimp1) embryo is indistinguishable from wild-type. S7 denotes somite 7. (C) 24 hpf Tg(hs::blimp1) embryo that received a heat pulse at 14 hpf showing supernumerary slow muscle fibers (cf. panel B). In panels B and C, Slow MyHC is shown in green and Prox1 in red. (D) Expression of smyhc1 mRNA (green) and fast myosin light chain protein (red) in an uninduced Tg(hs::blimp1) embryo. (E) Induction of ectopic Blimp1 expression in Tg(hs::blimp1) embryos at 14 hpf results in an increase in the number of slow muscle fibers (smyhc1 expression, green) at the expense of fast muscle fibers (red). In panels D and E, nuclear staining with DAPI is shown in blue and position of the notochord marked with “N”. (F) Late induction of the blimp1 transgene (at 18 hpf) did not result in significant differences in the number of slow fibers (slow MyHC, green; Prox1, red) compared to an untreated embryo (cf. panel B). (G) The slow muscle marker Prox1 (red, arrows) is ectopically induced in multi-nucleated fast fibers (expressing fast myosin light chain, cyan) by induction of the blimp1 transgene at 18 hpf. (H–J) Numbers of slow muscle fibers in somites 5–11 of Tg(hs::blimp1) embryos were scored and presented as bar charts for blimp1 induction at 10 hpf (H), 14 hpf (I) and 18 hpf (J). There are significant differences in the number of slow fibers in somites 5–7 with induction of blimp1 at 10 hpf (p < 0.05, n = 12). More myoblasts were responsive to Blimp1 activity at 14 hpf resulting in a more significant increase in the number of slow fibers (p < 0.01, n = 12). Induction of blimp1 at 18 hpf did not result in significant changes in the number of slow fibers (n = 15). The error bars represent the standard error of mean (S.E.M), ** represents p < 0.01 and * represents p < 0.05). Panels A–C, F, and G depict lateral views; panels D and E depict transverse sections. Scale bars = 25 μm. The scale bar in A applies to B, C, F and G and the scale bar in D applies to E. EXPRESSION / LABELING:
|
|
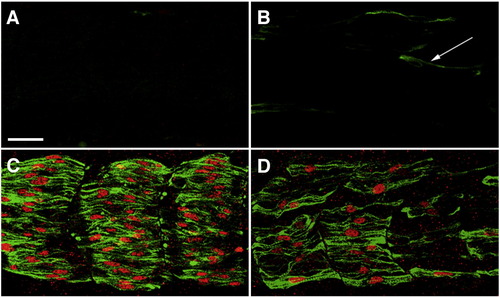
Mammalian Blimp1 protein can rescue slow myogenesis in zebrafish embryos that lack endogenous Blimp1 activity. (A) A smo embryo showing lack all slow muscle fibers. (B) An ubo embryo with a few residual slow MyHC-positive cells (green, arrow). These cells are not properly differentiated slow muscle fibers, as they do not express Prox1 (red). (C, D) Expression of mammalian Blimp1 from a heat-inducible promoter can rescue slow muscle development in both smo (C) and ubo embryos (D). All panels depict lateral views of 24 hpf embryos stained with mAb F59 (green) and anti-Prox1 (red). Scale bar = 25 μm. The scale bar in A applies to B–D. EXPRESSION / LABELING:
|
|
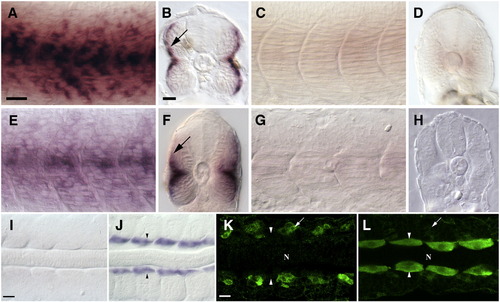
Loss of Blimp1 activity results in mis-regulation of slow as well as fast muscle genes in the precursors of the slow muscle lineage. (A, B) Expression of slow troponin-c in slow muscles of 24 hpf wild-type embryos. In B, staining in the superficial slow muscle layer is visible (arrow). (C, D) Expression of slow troponin-c is lost in 24 hpf ubo embryos. (E, F) Expression of slow myosin light chain in slow muscles of wild-type 24 hpf embryos. In (F) staining in the superficial slow muscle layer is visible (arrow). (G, H) Expression of slow myosin light chain is lost in 24 hpf ubo embryos. (I) mylz2 is not expressed in the adaxial cells of a 14 hpf wild-type embryo. (J) mylz2 is ectopically expressed in the adaxial cells of a blimp1 morphant embryo (arrowheads, n = 25/25). (K) Fast myosin light chain (green, detected with mAb F310) is normally expressed exclusively in fast muscle precursors in a 16 hpf wild-type embryo (arrow) and is absent from adaxial cells (arrowheads). (L) Fast myosin light chain protein is ectopically expressed at high levels in adaxial cells of a 16 hpf ubo embryo (arrowheads). Expression in fast muscle precursors is indicated (arrow). In K and L, “N” marks the position of the notochord. Panels A, C, E and G depict lateral views; panels B, D, F and H depict transverse sections; panels I–L show dorsal flat mounts. Scale bars = 25 μm. The scale bar in A is applicable for C, E and G, the scale bar in B applies to D, F and H. The scale bar in I is applicable in J and the scale bar in K is applicable in L. EXPRESSION / LABELING:
|
|
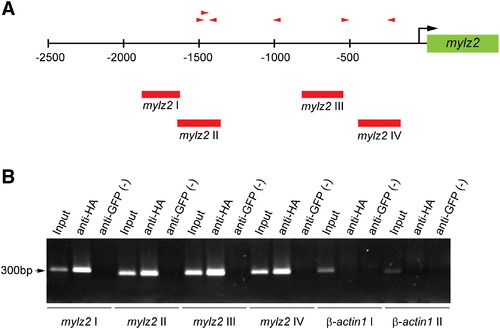
ChIP analysis reveals that Blimp1 binds to the promoter of the mylz2 gene. (A) The mylz2 gene promoter region contains six putative Blimp1 binding sites (red arrowheads, direction indicates orientation) based on variations of the mouse Blimp1 consensus binding sequence. Numbers indicate base pairs relative to the transcription start site (black arrow). Positions of PCR primers designed to test for Blimp1 binding are indicated (red bars, mylz2 I–IV). (B) PCR analysis of chromatin before ChIP (Input), and after immunoprecipitation with anti-HA antibodies or anti-GFP antibodies (negative control). Sequences near the β-actin1 gene served as a control for non-specific ChIP (β-actin I, II). |
Reprinted from Developmental Biology, 324(2), Liew, H.P., Choksi, S.P., Wong, K.N., and Roy, S., Specification of vertebrate slow-twitch muscle fiber fate by the transcriptional regulator Blimp1, 226-235, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.