- Title
-
An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10
- Authors
- Dutton, J.R., Antonellis, A., Carney, T.J., Rodrigues, F.S., Pavan, W.J., Ward, A., and Kelsh, R.N.
- Source
- Full text @ BMC Dev. Biol.
|
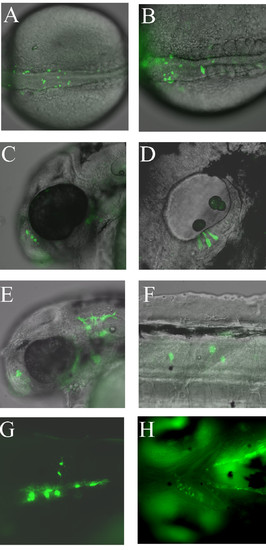
Examples of mosaic GFP expression from transient transgenic sox10:GFP zebrafish injected with psox10-1252:GFP plasmid at the 1-cell stage. A, B. Dorsal views of two separate examples of pre-migratory neural crest GFP expression at the 20-somite stage. C. GFP expression in nasal cells at 48 hpf. D. GFP expression in otic epithelium at 48 hpf. E. GFP expression in xanthophores at 48 hpf. F. GFP expression in interneurons at 48 hpf. G. GFP expression in oligodendrocytes in trunk spinal cord at 96 hpf. H. Jaw cartilage expression at 96 hpf. All panels contain fluorescent images from live embryos (overlaid on DIC images in A-F). Orientation is with anterior to the left, dorsal uppermost except A, B (dorsal) and H (ventral). |
|
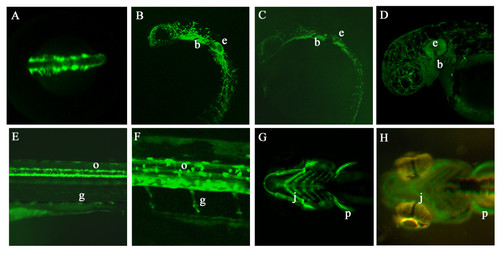
GFP expression in germline transgenic sox10:GFP embryos. A. Dorsal view of head and anterior trunk of a Tg(-4725sox10:GFP)ba4 embryo at 16 hpf showing GFP expression in premigratory and early migrating neural crest cells. B, C. Lateral views of transgenic embryos from lines Tg(-4725sox10:GFP)ba4 (B) and Tg(-1252sox10:GFP)ba5 (C) to show similarities in GFP expression pattern at 24 hpf. (e ear, b branchial arches). D. Lateral view of head of a Tg(-4725sox10:GFP)ba4 embryo at 48 hpf showing extensive GFP expression throughout cranial pigment cells, ear (e) and branchial arches (b). E, F. Lateral views of the trunk of Tg(-4725sox10:GFP)ba4 embryos expressing GFP in oligodendrocytes (arrow) and Schwann cells (arrowhead). G, H. Ventral views comparing GFP expression in the jaw cartilage (j) and pectoral fins (p) of transgenic embryos from Tg(-4725sox10:GFP)ba4 (F) and Tg(-1252sox10:GFP)ba5 (G) at 96 hpf. EXPRESSION / LABELING:
|
|
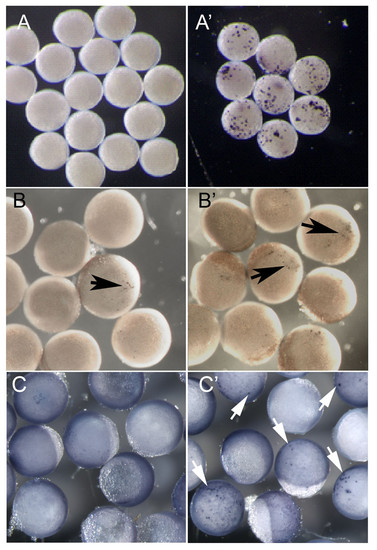
Functional testing of transcription factors implicated in sox10 regulation. A, B. Early zebrafish embryos (6 hpf), injected with plasmid constructs at the 1-cell stage, were divided and half subjected to a 60 minute 37°C heatshock. All were left to develop for a further 1 hour at 28.5°C before fixing. sox10 in situ hybridisation was performed to assess ectopic sox10 expression. A, A′ embryos injected with Hs:notchΔEmv. B, B′ embryos injected with Hs:Δβ catenin. C. Embryos of the transgenic line Tg(-4725sox10:GFP)ba3 were injected with equivalent constructs lacking GFP-coding regions. C, C′ embryos injected with hs:zsox9b. A-C no heatshock, A′-C′ with heatshock. Ectopic sox10 or gfp expression is indicated by the coloured reaction product (arrows). Note that the low level expression in B reflects the known leakiness of this heatshock promoter [71]. |
|
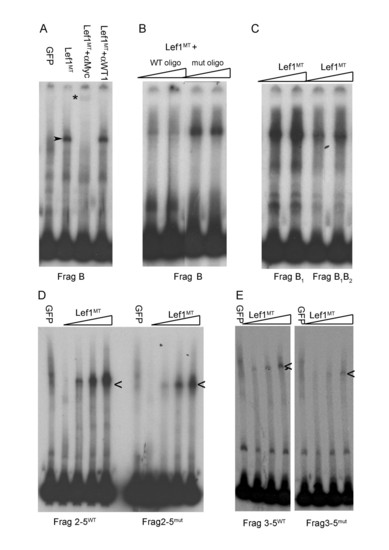
(A,) Electrophoretic mobility shift assays show specific Lef1 binding to 3′ sox10 intron 1 DNA sequences contained in Fragment (Frag) B. A specific band of reduced electrophoretic mobility (arrowhead) is observed when the 3′ intron 1 fragment B is incubated with in vitro made myc-tagged Lef1 (Lef1MT) protein but not when incubated with in vitro made GFP protein. The specific Lef1MT/DNA complex is lost in the presence of anti-myc antibody and replaced by a supershifted band (*); this band is specific to Lef1MT since this supershift is not seen with an unrelated (anti-WT1) antibody. (B) Competition with excess unlabeled oligonucleotide containing a wild type Lef1 binding site (WT oligo) abolishes the specific binding of Lef1 with Fragment B but oligonucleotide in which the Lef1-binding motif is mutated (mut oligo) is unable to compete for specific binding. (C) Site directed mutagenesis of the conserved Lef1 binding site B1 has little effect on Lef1 binding to Fragment B, additional mutation of site B2 (to give Frag B1,B2) reduces binding of Lef1. (D) Lef1 binding to fragment 2–5 (2–5WT) is reduced by mutagenesis of sites B1, B2 and site 5 in 2–5mut. (E). Reduced Lef1 binding to fragment 3–5 containing only two conserved Lef1 binding sequences. |
|
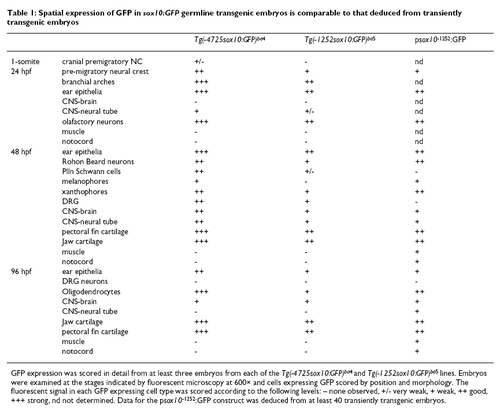
Spatial expression of GFP in sox10:GFP germline transgenic embryos is comparable to that deduced from transiently transgenic embryos EXPRESSION / LABELING:
|