- Title
-
V2a and V2b neurons are generated by the final divisions of pair-producing progenitors in the zebrafish spinal cord
- Authors
- Kimura, Y., Satou, C., and Higashijima, S.I.
- Source
- Full text @ Development
|
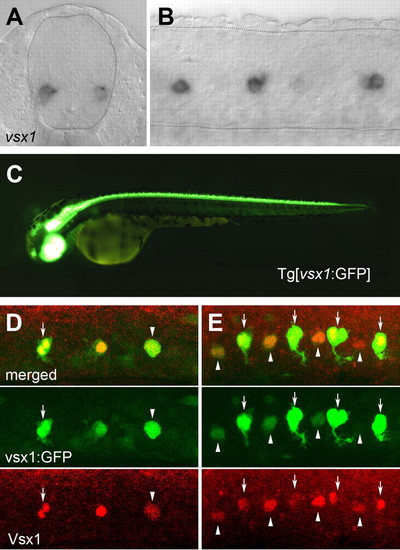
vsx1 mRNA and vsx1:GFP expression in spinal cord. (A,B) vsx1 mRNA expression in 22 hpf embryos. Broken lines outline the boundary of the spinal cord. (A) Cross-section. (B) Lateral view of a whole mount. (C) Tg[vsx1:GFP] fish at 2.5 dpf. (D,E) Expression of GFP and Vsx1 in Tg[vsx1:GFP] embryos at (D) 17 hpf and (E) 19 hpf. Arrows indicate pairs of labeled cells. Arrowheads indicate single labeled cells. EXPRESSION / LABELING:
|
|
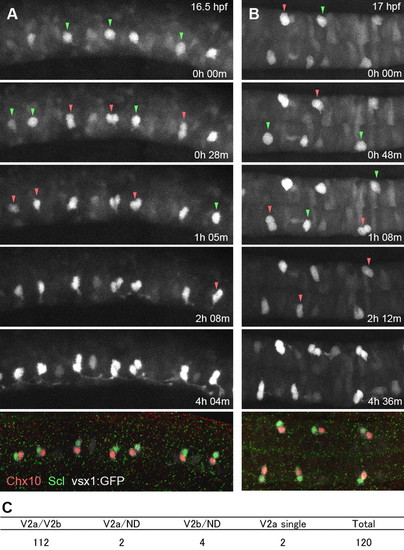
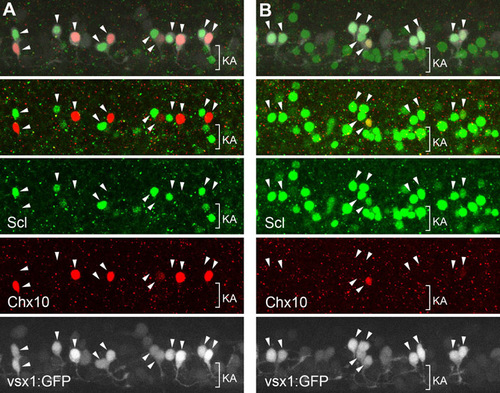
Time-lapse imaging of vsx1-GFP cells showing that V2a and V2b neurons are produced as siblings. (A,B) Time-lapse imaging of vsx1-GFP cells, followed by immunohistochemical localization of Chx10 (red, V2a) and Scl (green, V2b). (A) Lateral view. (B) Dorsal view. Images are montages of stacked optical sections. Green arrowheads indicate cells prior to division. Red arrowheads indicate paired cells after division. Movies for these analyses are available in the supplementary material. (C) Summary of time-lapse imaging. EXPRESSION / LABELING:
|
|
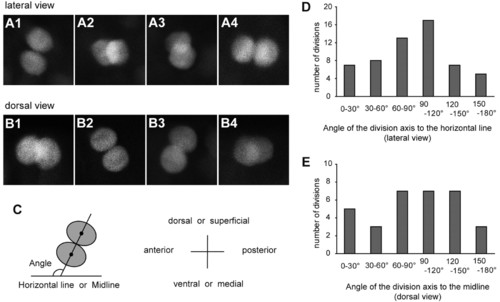
Division axes of p2 intermediate progenitors are not fixed. (A,B) Confocal stacked images of vsx1-GFP sibling cells immediately after division. Embryos were analyzed at around 16-17 hpf. (A1-A4) Lateral views. Dorsal is towards the top. (B1-B4) Dorsal views. Superficial is towards the top. (C) Schematic of the method for division axis measurements. For each division, a line connecting the centers of the two sister cell (projection of division axis) was drawn. The angle of this line to the horizontal line (lateral views) or to the midline (dorsal views) was measured. (D,E) Quantification of the division angle. EXPRESSION / LABELING:
|
|
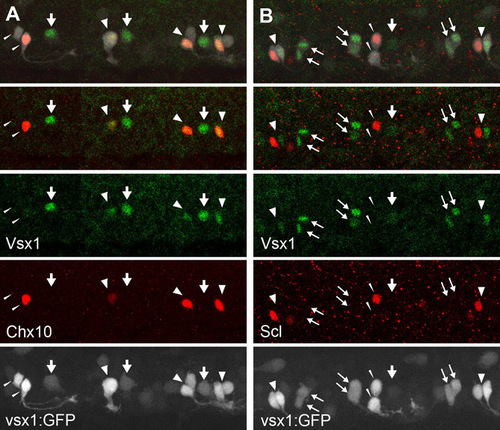
Vsx1 protein expression in the V2 neuron lineage. (A) Dual immunohistochemical staining of Vsx1 (green) and Chx10 (red) in a Tgvsx1:GFP (GFP signal is white) embryo at 20 hpf. (B) Dual immunohistochemical staining of Vsx1 (green) and Scl (red) in a Tgvsx1:GFP (GFP signal is white) embryo at 20 hpf. In both figures, the top panels show merged images of three channels (white, green and red), whereas the second panels from the top show merged images of two channels (green and red). Vsx1-expressing cells are either p2 intermediate progenitors or their progenies. For example, the single GFP cells marked by thick arrows probably correspond to p2 intermediate progenitors. Vsx1-expressing cells are also present as pairs (thin arrows). Each pair probably represents a sibling, and thus both of the progeny can express Vsx1. Often, however, Vsx1 expression is only present in one cell of the putative pair (fat arrowheads). In these cases, Vsx1-positive cells often co-express Chx10 (fat arrowheads in A), whereas Vsx1-expressing cells and Scl-expressing cells tend to make up pairs (fat arrowheads in B). These observations strongly suggest that Vsx1 protein expression occurs in the following manner in the V2 neuron lineage. First, Vsx1 protein expression is upregulated in p2 intermediate progenitors. After division, both progeny temporarily express Vsx1. The cells that will become V2b neurons (Scl positive) then lose Vsx1 expression, whereas the cells that will become V2a neurons continue to express Vsx1. We have also observed a pair of cells without detectable levels of Vsx1 in either cell (slim arrowheads; note that one of the cells expresses either Chx10 or Scl). These observations suggest that V2a neurons ultimately lose Vsx1 protein expression. EXPRESSION / LABELING:
|
|
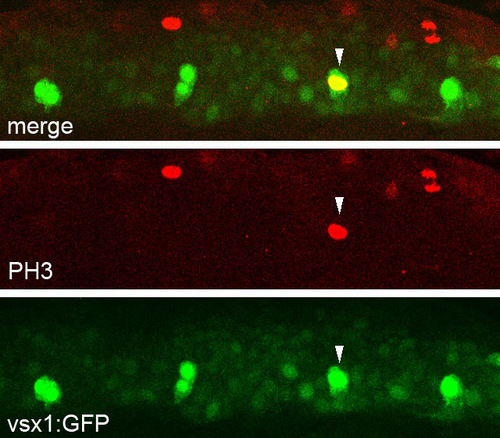
Mitotic cells are included among the vsx1-GFP cell population. Anti-PH3 (phospho-Histone H3) immunohistochemical staining in a Tgvsx1:GFP embryo at 17 hpf. A vsx1-GFP cell (arrowhead) is also positive for PH3. EXPRESSION / LABELING:
|
|
Notch-signaling affects the cell fate of V2a andV2b neurons. Double immunohistochemical staining of Chx10 (red) and Scl (green) in Tgvsx1:GFP embryos (GFP signal is white) at 24 hpf. (A) Control experiment (Tgvsx1:GFP alone). (B) Notch1a intracellular fragment (NICD, an activated form of Notch1a) was widely expressed in neurons following a cross between TgUAS:nic (Scheer et al., 2001) and TghuC:Gal4-VP16 (see Materials and methods), in a Tgvsx1:GFP background. In both figures, the arrowheads represent possible sibling vsx1-GFP cell pairs. In A, a Chx10-positive cell and a Scl-positive cell make up a pair. In B, both cells in a pair often express Scl but not Chx10. This observation suggests that the forced activation of Notch signaling produces greater numbers of V2b neurons at the expense of V2a neurons. Peng et al. (Peng et al., 2007) have previously presented similar results using gata3/scl and vsx1 as markers for V2b and V2a, respectively. It should be noted that, in addition to V2b neurons, Scl is also expressed in Kolmer-Agduhr (KA) neurons (denoted by KA in the figures). Developmentally, KA neurons derive from the pMN domain (KA′ neurons) and the p3 domain (KA″ neurons) (Park et al., 2004; Yeo and Chitnis, 2007; our unpublished observations). Because KA neurons are located in a medial region of the spinal cord, most are out of focus in A (and in other figures presented in this study). Consequently, only a small number of these neurons are visible in A. In B, a large number of Scl-positive cells, which are likely to be KA neurons, are present in a ventral region of the spinal cord (note that the cells denoted as KA are negative for vsx1-GFP). These neurons probably originated through fate conversion from motoneurons, as Delta-Notch signaling is involved in fate specification between motoneurons and KA neurons (Shin et al., 2007). It should also be noted that, in TghuC:Gal4-VP16 embryos, the expression of Gal4-VP16 was probably not restricted to the postmitotic neurons, despite the fact that huC is expressed exclusively in postmitotic neurons. Our in situ hybridization analyses of TghuC:Gal4-VP16 embryos showed that Gal4-VP16 was expressed not only in postmitotic neurons, but also in neural precursor cells, possibly owing to an effect of transgene integration position in the transgenic line. Thus, it remains to be established whether NICD induction in postmitotic neurons by a Gal4-UAS system is sufficiently early enough to induce a neuronal fate change from V2a to V2b. |
|
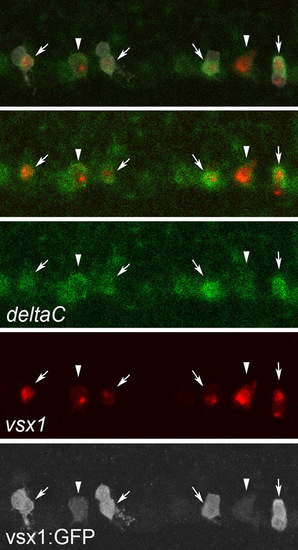
deltaC expression is upregulated in V2a neurons. Triple staining of deltaC mRNA (green), vsx1 mRNA (red) and GFP protein (white) in a Tgvsx1:GFP embryo at 19 hpf. The top panel shows a merged image of the three signals. The second panel from the top shows a merged image of the green and red channels. deltaC mRNA expression is present in p2 intermediate progenitors (arrowheads). In pairs of vsx1-GFP cells, strong deltaC expression is seen in only one of the cells (arrows). These cells also express vsx1. Thus, in a pair of vsx1-GFP cells, deltaC is likely to be upregulated in a cell that will ultimately differentiate into a V2a neuron. EXPRESSION / LABELING:
|
|
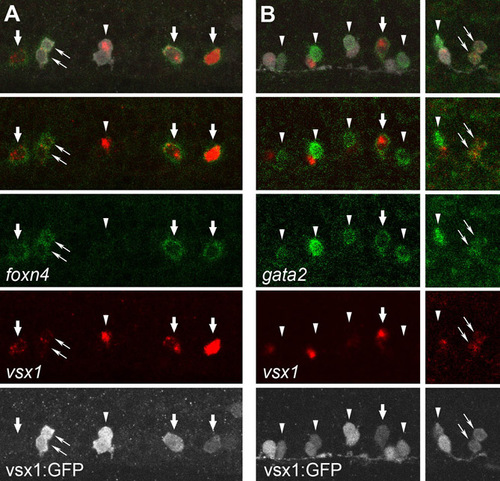
Expression profile of foxn4 and gata2 mRNA in the V2 neuron lineage. (A) Triple staining of foxn4 mRNA (green), vsx1 mRNA (red) and GFP protein (white) in a Tgvsx1:GFP embryo at 18 hpf. The top panel shows a merged image of three signals. The second panel from the top shows a merged image of the green and red channels. foxn4 is expressed in single vsx1-positive cells (thick arrows), which are likely to be p2 intermediate progenitors. The left-most cell that is positive for both foxn4 and vsx1 does not have detectable levels of GFP expression. The most likely explanation for this observation is that the cell is a very early stage p2 intermediate progenitor (accumulation of GFP has not yet reached at detectable level). foxn4-expressing cells can also be detected in a pair of vsx1-GFP cells (thin arrows), which are probably the progeny of a single progenitor. However, foxn4 expression can also be absent in both cells of a vsx1-GFP pair (arrowhead). This observation suggests that both of the cells have lost foxn4 expression. Thus, foxn4 mRNA expression in the V2 neuron lineage may proceed as follows: foxn4 expression is first upregulated in a p2 intermediate progenitor cell. After division, foxn4 is transiently present in both of the progeny. Subsequently, both cells lose foxn4 expression. It should be noted that the disappearance of foxn4 in the V2 progeny occurs earlier than that of vsx1. In the pair of cells marked by the arrowhead, vsx1 mRNA is still present in one of the cells (the vsx1-positive cell would probably differentiate into V2a neuron; see Fig. S1. (B) Triple staining of gata2 mRNA (green), vsx1 mRNA (red) and GFP protein (white) in a Tgvsx1:GFP embryo at 20 hpf. gata2 expression is expressed in single vsx1-positive cells (thick arrow) or in pairs of vsx1-GFP cells (thin arrows). In many of the paired cells, gata2 expression is detected only in one of the paired cells (arrowheads). gata2-negative vsx1-GFP cells usually express vsx1. Based on these observations, gata2 mRNA expression in the V2 neuron lineage may occur as follows: gata2 expression is first upregulated in p2 intermediate progenitors and after division is temporarily present in both of the progenies. One of the cells then loses gata2 expression (but continues to be positive for vsx1), and will differentiate into a V2a neuron. The other cell that continues to express gata2 will differentiate into a V2b neuron. EXPRESSION / LABELING:
|
|
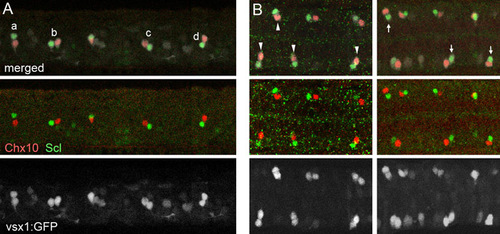
The final location of the V2a and V2b neurons is not fixed. Double immunohistochemical staining of Chx10 (red) and Scl (green) in Tgvsx1:GFP embryos (GFP signal is white) at 20 hpf. (A) Lateral view. (B) Dorsal view. V2a/V2b pairs show various configurations. For example, in the lateral view (A), the relative position of Scl-positive cells to their sibling Chx10-positive cells could be to the dorsal (a), to the anterior (b), to the posterior (c) and to the ventral (d). Similarly, in the dorsal view (B), the relative position of Scl-positive cells could be to the lateral (arrowheads) and to the medial (arrows). EXPRESSION / LABELING:
|