- Title
-
barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches
- Authors
- Sperber, S.M., and Dawid, I.B.
- Source
- Full text @ Dev. Biol.
|
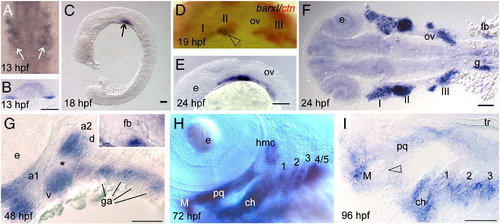
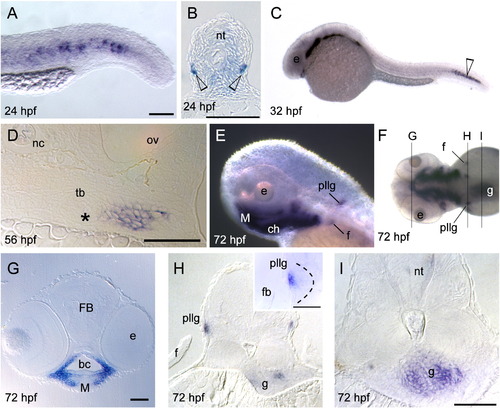
Barx1 expression. Embryos at (A, B) 13 hpf, (C)18 hpf, (D) 19 hpf, (E, F) 24 hpf, (G) 48 hpf, (H) 72 hpf, and (I) 96 hpf, were examined by in situ hybridization. (A) barx1 expression in the migrating neural crest in bilateral domains adjacent to the hindbrain (arrows). Anterior is at the top. (B) Coronal section through the hindbrain. (C) Lateral view of barx1 expression (arrow). (D) Double in situ of barx1 (dark blue, arrowhead) and crestin (red) co-expression in the neural crest streams indicated by Roman numerals. (E, F) Expression in the cranial neural crest streams. (G, G inset) barx1 expression in the pharyngeal arches and fin bud; a1, first arch; a2, second arch; d, dorsal expression domain; ga, gill arches; v, ventral expression domain; asterisk indicates the a2 intermediate region. (H) Lateral oblique view of barx1 expression in the chondrocytes. (I) Parasagittal section, open arrowhead indicates jaw joint. ch, ceratohyal; ceratobranchials are numbered; e, eye; fb, fin bud; g, gut primordium; hmc, hyomandibular condensation; M, Meckel's cartilage; ov, otic vesicle; pq, palatoquadrate; tr, trabecula. Scale bar: (B and I) 50 μm; (C, E–G) 100 μm. EXPRESSION / LABELING:
|
|
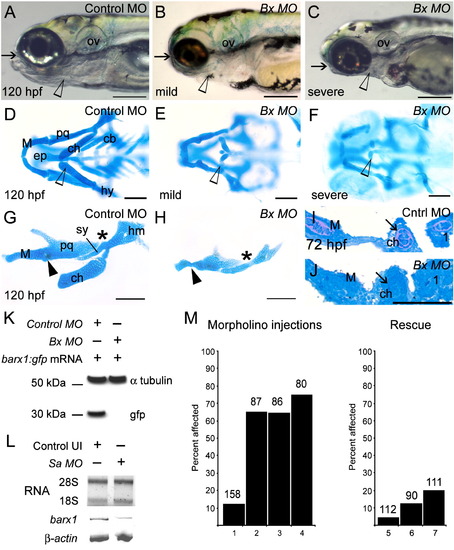
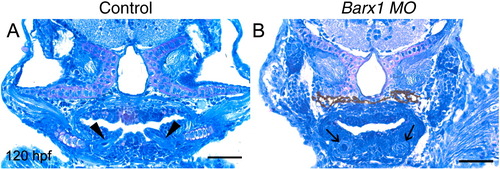
Attenuation of barx1 expression perturbs viscerocranial morphology and cartilage patterning. Larvae injected with (A) control MO (4 ng), and (B and C) Bx MO (4 ng). Alcian Blue staining of control MO (D) and Bx MO injected (E and F) 120 hpf larvae. Dissection of the first and second arches of control (G) and mildly affected larvae (H). (I and J) Parasagittal sections of 72 hpf embryos. (J) Bx MO (4 ng) injected compared with (I) control MO injected (4 ng). Anterior is to the left; (A–C, G–J) lateral views, (D–F) ventral view. (A–C) Arrows indicate mouth, and arrowheads the pharyngeal arches; (D–F) open arrowheads indicate the ceratohyal cartilage; (G, H) asterisk, perturbed fusion; black arrowhead, joint fusion between Meckel's cartilage (M) and the palatoquadrate (pq); cb, ceratobranchials; ch, ceratohyal; ep, ethmoid plate; hm, hyomandibular; ov, otic vesicle; sy, symplectic element. Scale bars: (A–C) 250 μm; (D–H) 100 μm; (J) 50 μm. (K, L) Effectiveness and specificity of the barx1 (Bx) and splice-acceptor (Sa) morpholinos. (K) Western analysis of in vivo attenuation of barx1:GFP translation in 24 hpf zebrafish embryos. (L) Total RNA extracts (18S and 28S bands shown) used for RT-PCR of barx1. β-actin used as a control. (M) Percentage of affected embryos comparing control MO, Bx MO and Sa MO, and rescue by co-injection of barx1 mRNA with a 5-bp mismatch to the ATG target site. Affected embryos are defined as those exhibiting micrognathia as a result of a reduction or loss of arch cartilage elements, as seen by Alcian Blue staining at 120 hpf. (1) Control MO 4 ng; (2) Bx MO 4 ng; (3) Sa MO 4 ng; (4) Bx MO 2 ng + Sa MO 2 ng. Rescue: (5) gfp mRNA 25 pg; (6) barx1 mRNA 25 pg; (7) Bx MO + Sa MO 2 ng each + barx1 mRNA 25 pg. Number of embryos indicated on top of each bar. PHENOTYPE:
|
|
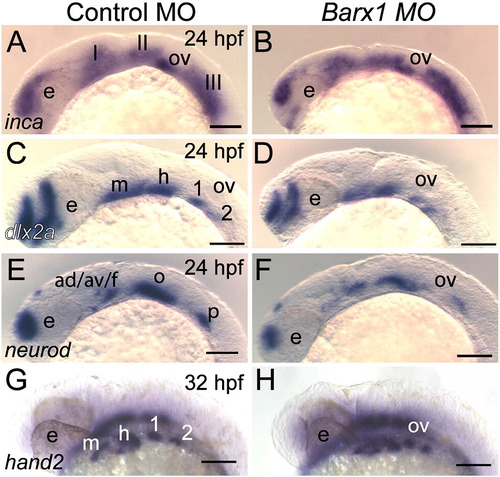
Initial patterning of the arches is maintained in barx1 morphants. (A, C, E and G) Control MO-injected embryos, (B, D, F and H) embryos co-injected with Bx MO + Sa MO, 2 ng each. Neural crest expression of inca (A, B) and dlx2a (C and D) at 24 hpf. (E, F) neurod expression at 24 hpf in neural ganglia precursors; (G and H) hand2 expression in ventral endoderm at 32 hpf. Neural crest streams marked in Roman numerals, gill arches indicated by numbers; e, eye; m, mandibular arch; h, hyoid arch; ov, otic vesicle; ad/av/f, anterodorsal/anteroventral lateral line/facial placode/ganglia; o, octavel/statoacoustic ganglia precursors; p, posterior lateral line placode/ganglion. Scale bar: 100 μm. |
|
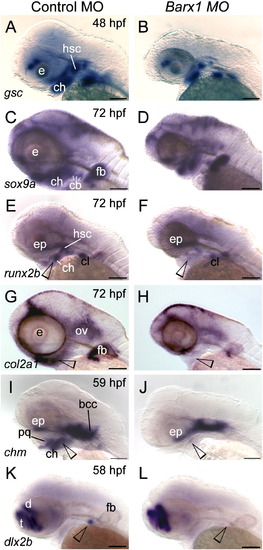
Barx1 is necessary for expression of chondrogenic markers within the pharyngeal arches. (A, C, E, G, I and K) Control embryos; (B, D, F, H, J and L) barx1 morphant embryos (see Fig. 4). Whole-mount in situ hybridization for (A, B) goosecoid (gsc), (C, D) sox9a, (E, F) runx2b, (G, H) type II collagen (col2a1), (I, J) chondromodulin (chm), and (K, L) dlx2b. Anterior is to the left; open arrowheads indicate sites of altered marker expression in the morphants; bcc, basicranial commissures; cb, ceratobranchials; ch, ceratohyal; cl, cleithrum; d, diencephalon; ep, ethmoid plate; fb, fin bud; hsc, hyosymplectic condensation; ov, otic vesicle; pq, palatoquadrate; t, telencephalon. Scale bar: 100 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
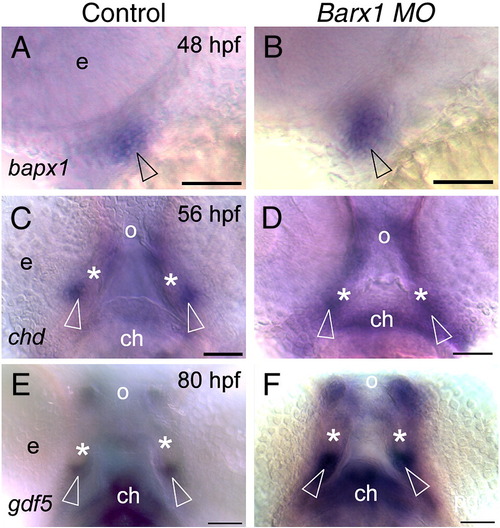
Loss of barx1 enhances chordin and gdf5 expression in the jaw joint. Whole-mount in situ hybridization for (A, B) bapx1, (C, D) chordin (chd), and (E, F) gdf5. (A, C, E) Control uninjected embryos; (B, D, F) barx1 morphants (see Fig. 4). Embryonic ages as indicated. (A, B) Lateral view, anterior to the left. (C–F) Ventral view, anterior at the top; asterisk indicates Meckel's cartilage, open arrowheads indicate jaw joint expression domains; e, eye; ch, ceratohyal; o orifice. Scale bar: (A–F) 50 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
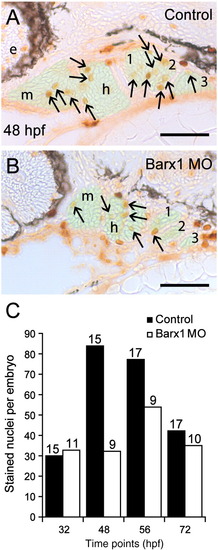
Barx1 positively influences cellular proliferation within the pharyngeal arches. (A, B) Sagittal serial sections (10 μm) of the pharyngeal arch region stained with Phosphohistone-H3 antibody. (A) Control embryo at 48 hpf. (B) barx1 morphant. The neural crest derived mesenchyme is overlaid in green; arrows indicate stained nuclei; e, eye; m, mandibular arch; h, hyoid arch; gill arches are numbered. Anterior is to the left. Scale bar: 50 μm. (C) Averaged cell proliferation in control uninjected embryos (black bars) and barx1 morphants (see Fig. 4; white bars) was determined within the mesenchyme as shown in panels A, B at the indicated four time points. Approximately fifty serial sections per embryo were examined. Number of embryos examined is indicated above each bar. Scale bar: (A, B) 50 μm. PHENOTYPE:
|
|
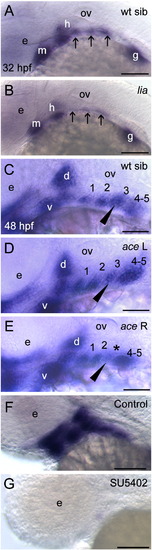
Barx1 expression in fgf mutant embryos. (A, B) Wild-type sibling and lia/fgf3 mutant. Arrows indicate sites of barx1 expression in the gill arches. (C–E) Wild-type sibling and left and right views of an ace/fgf8 mutant embryo. Left/right asymmetric expression indicated in the mutant (arrowhead); asterisk indicates reduced expression in the third gill arch. (F, G) Overstained control (DMSO) and embryo treated with the FGF receptor inhibitor SU5402 from 24 hpf to 48 hpf. Lateral views with anterior to the left, panel D was flipped for comparison; d, dorsal and v, ventral expression domains of the second arch; e, eye; gill arches are numbered; g, gut primordium; h, hyoid arch; m, mandibular arch; ov, otic vesicle. Scale bar: (A–G) 50 μm. |
|
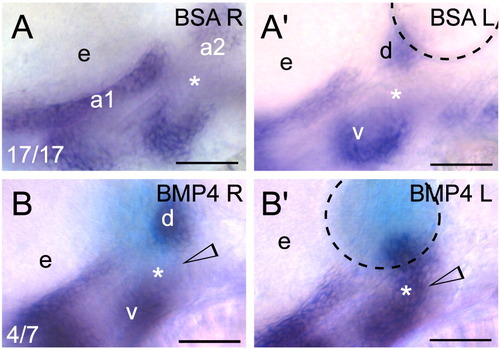
Exogenous BMP signaling influences barx1 expression within the zebrafish pharyngeal arches. (A, B) Whole-mount in situ hybridization of barx1 in the left (bead implanted) and right (control) side of single embryos. Right-side images were flipped for easier comparison. (A, A′) Control BSA white bead, (B, B′) BMP4 blue bead. Anterior is to the left; d and v, dorsal and ventral expression domains in the second arch; e, eye. Arrowhead indicates sites of barx1 misexpression; asterisk indicates intermediate region, normally devoid of barx1 expression at this stage. Number of affected embryos over total embryos with successful bead implantation is indicated in the panels A, B. Black dashed line outlines the bead. Scale bar: 50 μm. |
|
Whole-mount in situ hybridization of barx1 expression. (A–B) 24 hpf, (C) 32 hpf, (D) 56 hpf, and (E–I) 72 hpf. (A) Tail expression. (B) Cross-section of the trunk, open arrowheads indicates barx1 expression. (D) Coronal section of the fifth branchial arch adjacent to the tooth bud primordium (asterisk). (E) Lateral view of pharyngeal arch expression, posterior lateral line ganglia (pllg), and fin (f) expression domains. (F) Dorsal view indicating coronal serial sections in panels (G, H and I). (G) Expression in the arch mesenchyme of the jaw. (H) Cross-section through the pllg. (H inset) Higher magnification of expression in the posterior lateral line ganglia; dotted line outlines ganglion. (I) Cross-section through the gut wall (g). f, fin; FB, forebrain; M, Meckel's cartilage; nc, notochord; nt, neural tube; ov, otic vesicle; tb, tooth bud. Scale bar: (A) 100 μm, (B, D, G, H inset and I) 50 μm. EXPRESSION / LABELING:
|
|
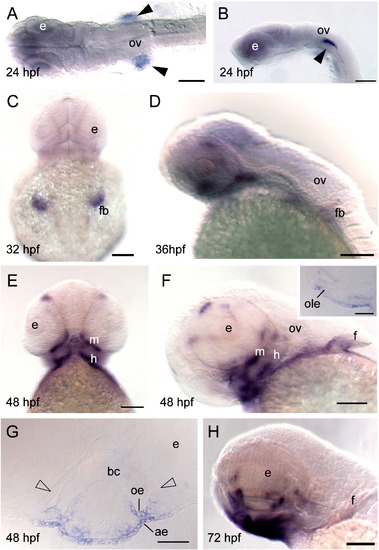
Whole-mount in situ hybridization of barx2 expression. (A) Dorsal view, anterior is to the left. (B, D, H) Lateral views, anterior to the left, (C) dorsal view; (E) ventral view; (C and E) anterior is to the top. (F) Lateral oblique view. (F inset) Sagittal section. (G) Coronal section. (A, B) Black arrowheads indicate bilateral sites of expression in the proximal aspect of the fin bud primordia. ae, aboral epithelium; bc, buccal cavity; e, eye; f, fin; fb, fin bud; h, hyoid arch; oe, oral epithelium; ole, olfactory bulb epithelium; ov, otic vesicle; m, mandibular arch. Scale bar: (A–F and H) 100 μm; (F inset and G) 50 μm. EXPRESSION / LABELING:
|
|
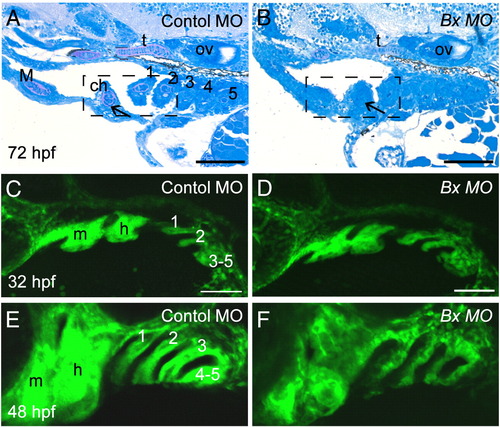
Perturbation of pharyngeal arch outgrowth and chondrocyte condensation in barx1 morphants. (A, C and E) Control MO-injected embryos, (B, D and F) barx1 morphants (see Fig. 4). (A, B) Pharyngeal arch sagittal sections at 72 hpf (see Figs. 3I and J). Arrow indicates the extracellular matrix surrounding the stacked chondrocytes. (C–F) fli1:GFP transgenic line at (C, D) 32 hpf and (E, F) 48 hpf. ch, ceratohyal cartilage; M, Meckel's cartilage; m, mandibular arch; gill arches are numbered; h, hyoid arch; ov, otic vesicle. Anterior is to the left in all panels. Scale bars: (A–F) 100 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Perturbation of tooth development in barx1 morphants. (A) Control larvae. (B) barx1 morphant (see Fig. 4). Coronal sections at 120 hpf. Arrowheads indicate erupting pharyngeal teeth; arrows indicate early cytodifferentiation stage tooth germs. Scale bar: (A, B) 50 μm. PHENOTYPE:
|

Unillustrated author statements PHENOTYPE:
|
Reprinted from Developmental Biology, 321(1), Sperber, S.M., and Dawid, I.B., barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches, 101-110, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.