- Title
-
Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease
- Authors
- Hirata, H., Watanabe, T., Hatakeyama, J., Sprague, S.M., Saint-Amant, L., Nagashima, A., Cui, W.W., Zhou, W., and Kuwada, J.Y.
- Source
- Full text @ Development
|
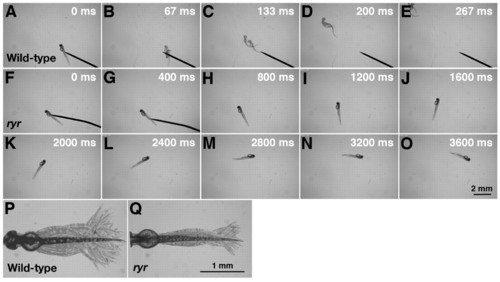
ryr mutant embryos exhibit slow swimming and weak muscle contractions in response to touch. (A-E) Mechanosensory stimulation induced a wild-type embryo (48 hpf) to swim away rapidly. (F-O) Touch induced an ryr mutant embryo (48 hpf) to swim, but only slowly. (P) Superimposition of all the frames from a video showing normal displacement of the trunk and tail during a swimming episode induced by tactile stimulation of a wild-type sibling. (Q) Displacement of the trunk and tail during swimming in ryr mutants is smaller compared with that of wild-type siblings. For P and Q, the heads of embryos were pinned on a Sylgard dish leaving most of the trunk and tail free. PHENOTYPE:
|
|
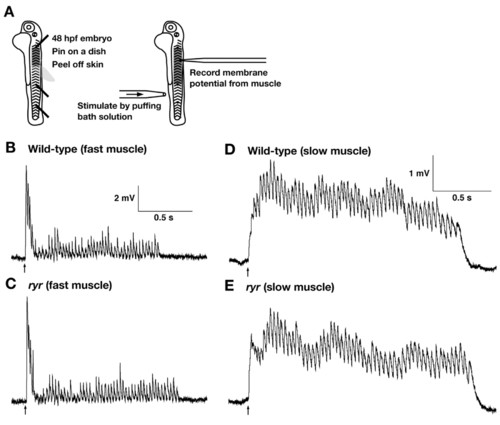
The CNS and NMJ function normally in ryr mutants. (A) Schematic summary of the experimental procedure. Embryos (48 hpf) are pinned on a dish with tungsten wires through the notochord so as not to damage the CNS. The skin is peeled off to allow access to the muscle cells. Muscle voltage responses are evoked by mechanosensory stimulation delivered by a puff of bath solution to the tail and are recorded with a patch electrode. (B,C) Voltage recordings from fast muscle fibers of a wild-type sibling (B) and an ryr mutant (C) displayed similar patterns of depolarizations (fictive swimming). (D,E) Voltage recordings from slow muscle of a wild-type sibling (D) and an ryr mutant (E) display similar rhythmic depolarizations. Arrows indicate the time of stimulation. |
|
Ca2+ transients are smaller in ryr mutant fast muscles. (A) Schematic summary of the experimental procedure. Calcium Green-1 dextran was injected into one cell of 8- to 16-cell-stage embryos. Embryos (48 hpf) were pinned on a dish and given mechanosensory stimulation by a puff of liquid to the tail. The relative level of Calcium Green-1 fluorescence ( PHENOTYPE:
|
|
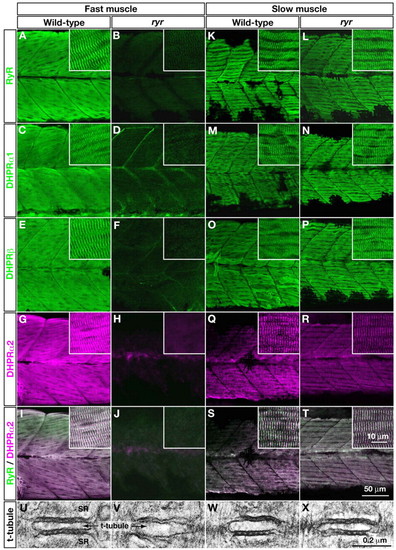
E-C-coupling components are dramatically decreased in ryr fast muscles. (A-H,K-R) The distribution of RyRs and DHPRs were assayed in fast and slow muscles at 48 hpf with anti-RyR (A,B,K,L), anti-DHPRα1 (C,D,M,N), anti-DHPRß (E,F,O,P) and anti-α2 (G,H,Q,R). Wild-type fast muscles express RyR (A), DHPRα1 (C), DHPRß (E) and DHPRα2 (G) in a striated pattern that presumably corresponds to the t-tubule-SR junctions, whereas clustering is not observed in mutant fast muscles (B,D,F,H). Wild-type slow muscles express RyR (K), DHPRα1 (M), DHPRß (O) and DHPRα2 (Q) in a striated pattern, as in wild-type fast muscles. ryr mutant slow muscles also express RyR (L), DHPRα1 (N), DHPRß (P) and DHPRα2 (R) in a striated pattern. (I,J,S,T) RyR (green) and DHPRα2 (purple) are colocalized in wild-type fast (I) and slow (S) muscle and in mutant slow muscle (T) but not in ryr mutant fast muscle (J). Insets show a higher magnification of muscle fibers. (U-X) Electron micrographs of t-tubule-SR junctions in muscles at 48 hpf. Putative RyR-DHPR aggregates are visible as dense particles between t-tubule and SR membranes in wild-type fast muscle (U) but not in ryr mutant fast muscle (V). The RyR-DHPR aggregates are present in both wild-type (W) and mutant (X) slow muscles. SR, sarcoplasmic reticulum. PHENOTYPE:
|
|
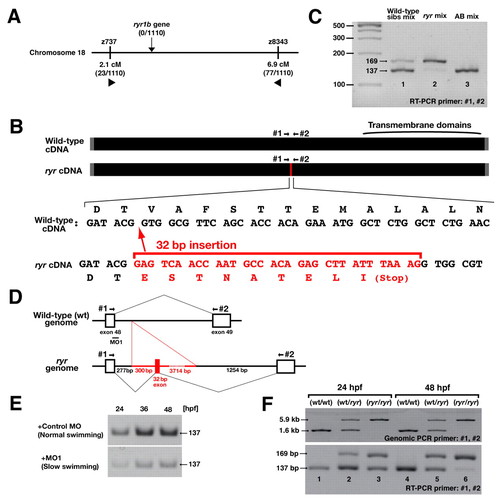
ryr mutant embryos have a mutation in ryr1b due to a DNA insertion that leads to aberrant splicing of ryr1b mRNA. (A) Meiotic mapping placed the ryr locus between z737 and z8343 in chromosome 18. No recombination was found with a polymorphic marker in the ryr1b gene (0/1110). (B) ryr mutants contained a 32-bp insertion (red) in ryr1b mRNA that added a nonsense codon 5′ to the transmembrane domains. (C) Reverse transcriptase (RT)-PCR using primers #1 and #2 (see B) amplified both a longer mutant band (169 bp) and a shorter wild-type band (137 bp) from a group of wild-type siblings (lane 1), whereas the mutant band was predominant in ryr mutants (lane 2). The wild-type band can be seen faintly in the mutant lane. Only the shorter band was amplified from wild-type AB embryos (lane 3). (D) ryr mutants carry a 4046-bp insertion (red) in the intron between exons 48 and 49 of ryr1b that includes the additional 32-bp sequence found in the mutant cDNA. (E) Injection of an antisense morpholino (MO1, see D) into recently fertilized wild-type embryos effectively decreases normal ryr1b mRNA at 24, 36 and 48 hpf, whereas control MO does not. A fragment of normal ryr1b mRNA was examined by RT-PCR with primers #1 and #2 (see D). (F) Genomic PCR and RT-PCR with primers #1 and #2 using individual embryos as a template. Genotypes, as shown, were identified by genomic PCR. Notice that, in mutants, the intensity of the wild-type (short) band amplified by RT-PCR was comparable to that of the mutant (long) band at 24 hpf (lane 3) but was diminished at 48 hpf (lane 6). |
|
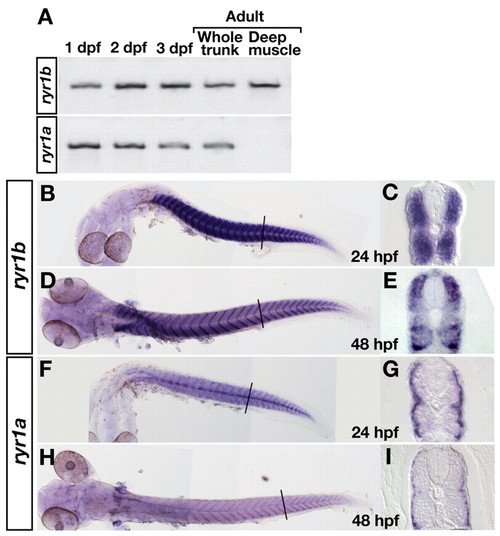
ryr1b and ryr1a are expressed by fast and slow muscle, respectively. (A) Reverse transcriptase (RT)-PCR shows that both ryr1b and ryr1a are expressed at 1, 2 and 3 dpf, and in adults. RT-PCR with mRNA from deep muscles indicates that ryr1b but not ryr1a is expressed by adult fast muscles. (B-I) In situ hybridization with ryr1b (B-E) and ryr1a (F-I) probes. Wholemounts show that both genes are expressed by muscle at 24 (B,F) and 48 (D,H) hpf. Cross-sections show that ryr1b is expressed by deep, fast muscles (C,E), whereas ryr1a is expressed by superficial, slow muscles but not by fast muscles (G,I). |
|
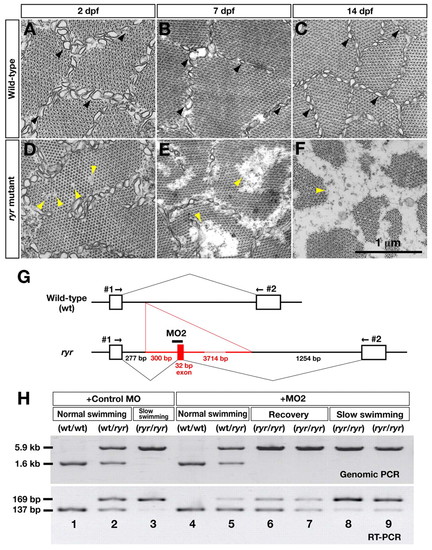
ryr mutants exhibit a minicore phenotype similar to that observed in MmD and the behavioral phenotype can be rescued by preventing aberrant splicing of ryr1b mRNA. (A-C) Electron micrographs of wild-type fast muscles showing sarcoplasmic reticulum (SR; black arrowheads), and bundles of actin and myosin at 2, 7 and 14 dpf. (D-F) Amorphous cores are seen in ryr mutants (yellow arrowheads). Diameters of the cores are 50-100 nm, 50-500 nm and 100-800 nm at 2, 7 and 14 dpf, respectively. The SR is disorganized at 7 dpf and missing at 14 dpf. (G) The behavioral defects of ryr mutants are treatable by an antisense morpholino that blocks abnormal splicing of ryr1b mRNA. A diagram showing the wild-type and mutant ryr1b genes with the location of the morpholino (MO2) that was designed against the splice acceptor site of the aberrant exon of the mutant gene is shown. (H) Control MO (lanes 1-3) or MO2 (lanes 4-9) was injected into the progeny of an ryr carrier incross. Swimming of injected embryos was examined at 36 hpf, and the head and trunk of individual embryos were subjected to genomic PCR and RT-PCR, respectively. Control MO-injected wt/wt embryos exhibited normal swimming and expressed only the wild-type, short fragment by RT-PCR (lane 1). Control MO-injected wt/ryr embryos exhibited normal swimming and expressed both the wild-type short and the mutant long fragments (lane 2). Control MO-injected ryr/ryr embryos exhibited slow swimming and predominantly expressed the mutant fragment (lane 3). MO2-injected wt/wt embryos exhibited normal swimming and expressed only the wild-type, short fragment (lane 4). MO2-injected wt/ryr embryos exhibited normal swimming and expressed both fragments (lane 5). In mutant embryos (ryr/ryr) showing recovery of swimming, normal splicing (short band) was increased (lanes 6, 7 compared to lane 3). On the other hand, in mutants that exhibited slow swimming, the proportion of wild-type mRNA was much lower (lanes 8, 9) compared to mutants that exhibited recovery (lanes 6, 7). EXPRESSION / LABELING:
PHENOTYPE:
|

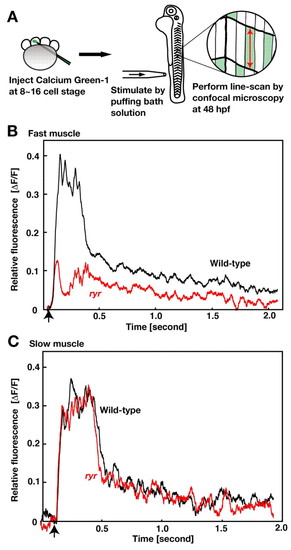
 F/F) in muscle fibers was assayed by line-scanning with a confocal microscopy. (B) The transient increase in relative fluorescence represents the transient increase of Ca2+ in the fast muscle following stimulation (arrow) of a wild-type embryo (black) and that of an ryr mutant (red). (C) The transient increase of Ca2+ in slow muscle is unperturbed in ryr mutants (red) compared to wild-type (black).
F/F) in muscle fibers was assayed by line-scanning with a confocal microscopy. (B) The transient increase in relative fluorescence represents the transient increase of Ca2+ in the fast muscle following stimulation (arrow) of a wild-type embryo (black) and that of an ryr mutant (red). (C) The transient increase of Ca2+ in slow muscle is unperturbed in ryr mutants (red) compared to wild-type (black).