- Title
-
Cohesin-dependent regulation of Runx genes
- Authors
- Horsfield, J.A., Anagnostou, S.H., Hu, J.K., Cho, K.H., Geisler, R., Lieschke, G., Crosier, K.E., and Crosier, P.S.
- Source
- Full text @ Development
|
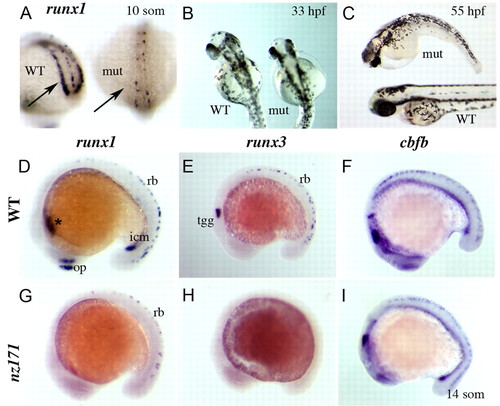
Zebrafish nz171 homozygous mutants lack runx1 and runx3 expression, but retain normal expression of cbfb. (A-C) runx1 expression and morphology of nz171 mutants. (A) Loss of runx1 expression in the PLM (posterior lateral-plate mesoderm; arrows) of an nz171 mutant embryo (mut) compared with wild-type sibling (WT); posterior views of 10-somite embryos. (B,C) Morphology of nz171 mutant (mut) embryos compared with siblings (WT) at the stages indicated (B, dorsal; C, anterior to the left). (D-I) Expression of runx1, runx3, and cbfb in wild-type and nz171 mutant embryos (14-somite embryos, anterior to the left). In nz171 mutants, expression of runx1 is lost in the ICM (intermediate cell mass; icm), ALM (anterior lateral-plate mesoderm; *) and olfactory placode (op) but retained in a subset of the Rohon-Beard (RB) mechanosensory neurons (rb) (G compared with D, wild type). nz171 mutants lack all runx3 expression (H compared with E, wild type). cbfb expression is normal in nz171 mutants (I compared with F, wild type). EXPRESSION / LABELING:
|
|
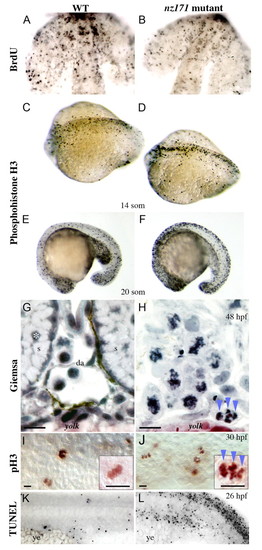
Cell cycle status of zebrafish nz171 mutant embryos compared with wild-type siblings. (A,B) BrdU-labeling to detect cells in S-phase in forebrain and eyes of flat-mounted 18-somite embryos, anterior to top. (C-F) Dorsal (C,D) and lateral (E,F) views of anti-phosphohistone H3 (pH3)-labeling to detect cells in M phase at the stages indicated (anterior to the left). (G-J) Disordered mitotic chromosomes in nz171 mutants. (G,H) Transverse sections bisecting the yolk extension of wild-type (G) and nz171 mutant (H) embryos stained with Giemsa (dorsal to top). Whereas recognizable structures (s, somite; da, dorsal aorta) and normal nuclei (example marked by asterisk) are visible in wild-type embryos, no such structures are visible in nz171 mutants, which have abnormal nuclei with condensed, disorganized chromosomes. pH3-labeled chromosomes of a similar configuration to H (blue arrowheads) appear in nz171 (J) but not in wild-type (I); lateral flat-mounted embryos, mid-trunk (high magnification inset). Scale bar: 20 µm. (K,L) TUNEL labeling indicates prevalent cell death in the ICM and neuronal regions of nz171 embryos; lateral views of tail regions, anterior to the left, showing the ICM region just above the yolk extension (ye). PHENOTYPE:
|
|
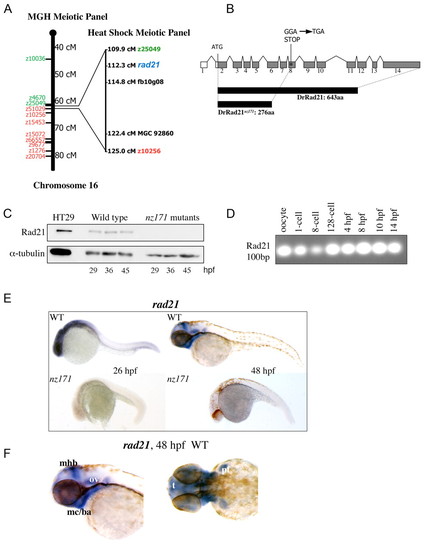
The nz171 mutation affects the zebrafish rad21 gene. (A) nz171 maps to a region on chromosome 16 containing rad21. Schematic of chromosome 16 from the centromere `southwards', cM (centiMorgans) indicated as per the MGH Meiotic Mapping Panel (http://zebrafish.mgh.harvard.edu/). SSLP markers determined to be `north' of the mutation are indicated in green and `south' markers in red. Right, expansion of the interval shown as mapped on the Heat Shock Meiotic panel (Woods et al., 2000) on which nearest north and south markers are shown. The rad21 gene is shown in blue. (B) Schematic representation of the rad21 gene, coding region shaded gray. A truncated protein is produced as a result of the Gly-277-STOP mutation in exon 8 (asterisk). (C) Rad21 protein is not present in nz171 homozygote embryos. Immunoblot showing Rad21 protein and α-tubulin loading control. Positive control is 5 µg protein isolated from HT29 colon carcinoma cells. (D-F) rad21 transcription in wild-type and nz171 embryos. (D) RT-PCR with rad21-specific primers indicates rad21 transcript is maternally inherited in zebrafish embryos. PCR products separated by agarose gel electrophoresis. (E,F) Expression pattern of rad21 in whole-mount wild-type and nz171 embryos at stages indicated (anterior to the left). rad21 transcript is undetectable in nz171 mutants (E, lower panels). (F) Lateral (left) and dorsal (right) views of anterior regions showing specific wild-type expression of rad21 (mhb, midbrain-hindbrain boundary; mc/ba, mandibular cartilage/branchial arches; ov, otic vesicle; t, telencephalon; pf, pectoral fin). |
|
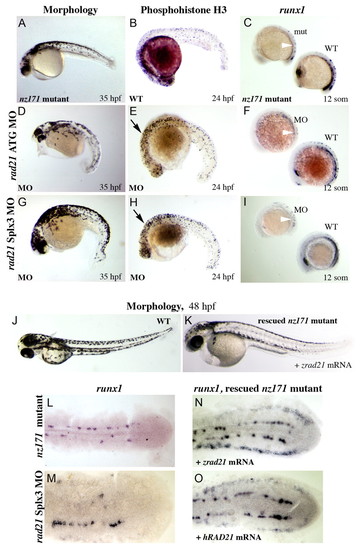
Morpholino knockdown of the rad21 gene phenocopies the nz171 mutation, both of which are rescued by injection of rad21 mRNA. (A-I) rad21 morphants phenocopy the nz171 mutation. (D,G) Embryos injected with antisense morpholino oligonucleotides targeting the start codon (rad21 ATG MO; D) and 5' donor site of exon 3 (rad21 Splx3 MO; G) physically resemble nz171 mutants (example in A) at 35 h.p.f. (B,E,H) pH3-labeled cells in rad21 morphants (E,H) compared with wild type (B). Morphants exhibit increased numbers of cells in M phase (black arrows). (C,F,I) runx1 expression in nz171 mutant (C) and rad21 morphant embryos (F,I), shown alongside wild type (WT, wild type; mut, nz171 mutant; MO, morphant). Reduced PLM runx1 expression is indicated by white arrowheads. All views lateral, anterior to the left. (J-O) The nz171 mutant phenotype is rescued by injection of a rad21 mRNA. (J,K) The morphology of nz171 mutants approaches wild type in rad21-injected embryos. (L-O) Expression of runx1 in the PLM of flat-mounted 10-somite embryos, anterior to the left, dorsal views. runx1 expression is absent from the PLM in nz171 (L) and virtually absent in a rad21 morphant (M). runx1 expression is rescued in the PLM after injection with zebrafish (N) or human (O) rad21 mRNA. See Fig. 5, Fig. 6A for wild-type expression of runx1. EXPRESSION / LABELING:
PHENOTYPE:
|
|
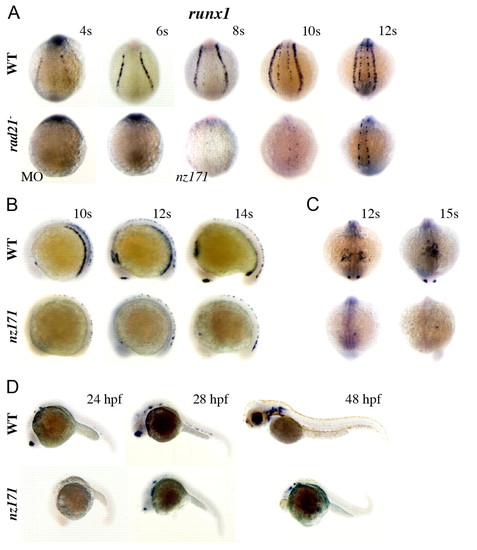
runx1 expression fails to initiate in the ALM and PLM of zebrafish embryos compromised for Rad21, but late neuronal runx1 expression is Rad21-independent. (A-D) Whole-mount embryos stained for runx1 expression. For all, upper panels are wild type, lower panels are rad21ATGMO morphants (MO) or nz171 homozygotes as indicated. (A) Posterior views of embryos, stages as indicated. rad21 morphants (MO; 0.5 pmol) at the 4-somite stage (4s; 14/15 embryos) and 6s (23/25 embryos) were runx1 negative. Expression of runx1 in RB cells is just detectable by the 8-somite stage in nz171 mutants, but PLM expression remains absent throughout. (B) Lateral views, stages as indicated. runx1 mRNA is not detected in the PLM or ICM, or in the olfactory placode of nz171 mutants, but is present in a population of RB cells. (C) Anterior views, stages as indicated. The occasional runx1-positive cell is visible in the ALM of nz171 mutants. (D) Lateral views, stages as indicated. At 24 h.p.f., nz171 homozygotes appear delayed and display no runx1 expression. By 28 h.p.f., neuronal and olfactory placode expression has initiated in mutants. Hematopoietic expression remains absent, c.f. wild type, where expression of runx1 is observed in the ventral wall of the dorsal aorta. Anterior is to the left for B and D. EXPRESSION / LABELING:
|
|
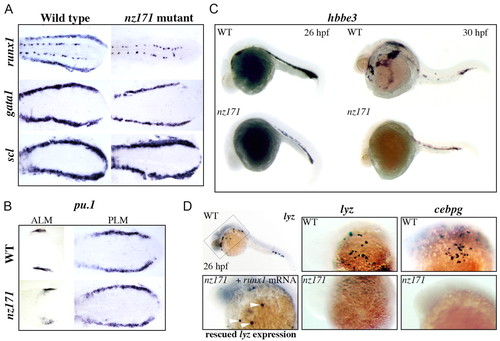
Expression of genes that mark `early' versus differentiated hematopoietic cells in zebrafish nz171 mutants, and partial rescue of differentiated cells by microinjection of runx1 mRNA. (A,B) Expression of runx1 and gata1 is altered in nz171 mutants. Flat preparations of 10-somite embryos, anterior to the left. Expression of runx1, gata1 and scl in the PLM (A), and pu.1 in the ALM and PLM (B) of wild-type siblings and nz171 mutants. gata1 expression in nz171 mutants is noticeably reduced compared with wild type. (C,D) Expression of genes that mark differentiated hematopoietic cells is reduced or absent in nz171 mutants. Whole-mount embryos, anterior to the left. (C) Reduced expression of hbbe3 (hemoglobin beta embryonic3) in nz171 mutant embryos (lower panels) compared with wild type (upper panels). (D) Loss of lyz (lysozyme) expression in nz171 is rescued by microinjection of runx1 mRNA. Upper left, whole-mount 26 h.p.f. embryo stained for lyz expression. The box indicates the region shown at higher magnification in the other panels. Upper panels, wild-type expression of lyz and cebpg in 26 h.p.f. embryos, as indicated. Lower center and right, similar views of 26 h.p.f. nz171 homozygotes, showing no expression of lyz and cebpg. Lower left panel, rescued expression of lyz in 26 h.p.f. embryos indicated by white arrowheads (21/23 mutant embryos rescued). EXPRESSION / LABELING:
|
|
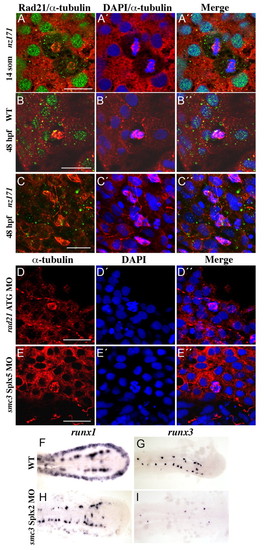
Zebrafish cohesin functions in sister chromatid cohesion and is necessary for normal expression of Runx genes in early embryogenesis. (A-C) Mitotic defects in nz171 embryos. Confocal immunofluorescence images, stained for Rad21 (green) and α-tubulin (red), with DNA stained with DAPI (blue), as indicated on top of the panels; embryo identity is on the left. By 48 h.p.f., Rad21 protein is absent from nz171 embryos, and multiple cells have arrested in mitosis (C-C''). (D,E) Knocking down the function of Rad21 (D-D'') or Smc3 (E-E'', exon 5 splice site) reproduces the mitotic phenotype observed in nz171 embryos at 48 h.p.f. Confocal immunofluorescence images, stained for α-tubulin (red), with DNA stained with DAPI (blue), as indicated in the panels (top); embryo identity is on the left. For A-E, images are of flat-mounted embryo tails; scale bar is 30 µm. (F-I) Loss of runx1 and runx3 expression in smc3 morphants. Flat-mounted 10-somite embryos, anterior to the left. smc3 morphants (exon 2 splice site) show loss of runx1 expression in the PLM (H), and runx3 expression in the RB cells (I). EXPRESSION / LABELING:
PHENOTYPE:
|
|
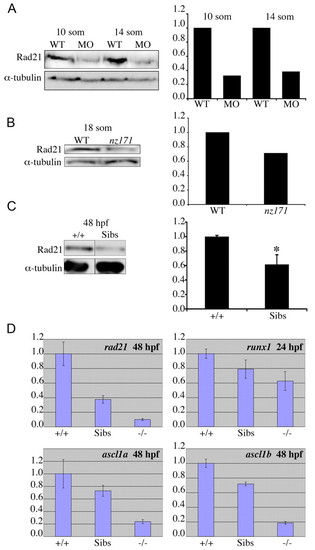
Maternally inherited Rad21 protein can sustain further cell division, but not gene expression; loss of one copy of rad21 affects transcription of runx1, ascl1a and ascl1b. (A,B) Rad21 protein is detectable in early rad21 morphant and mutant zebrafish embryos at stages when gene expression is compromised, but cell division can still occur. Protein was isolated from wild-type embryos and rad21ATGMO morphants (A) or nz171 homozygotes (B) at the stages indicated. Protein quantities were assessed by immunoblotting, with levels relative to α-tubulin. The graphs on the right show quantification of the immunoblots on the left. (C,D) Loss of one copy of rad21 reduces rad21 transcript and protein levels, and affects gene expression. (C) Protein was isolated from nz171 siblings (Sibs: includes +/+ and +/- embryos, which are phenotypically indistinguishable) and wild-type (+/+) controls at 48 h.p.f. Relative levels of Rad21 protein in nz171 siblings and wild-type embryos were quantified by immunoblotting and results are presented in the graph on the right (values represent the mean±s.e.m. of three independent experiments performed in triplicate. *P<0.01, Student′s t-test). A representative immunoblot appears on the left. (D) Quantitative RT-PCR of cDNA generated from pooled wild-type (+/+), nz171 sibling (Sibs) and nz171 mutant (-/-) embryos. Bar charts are representative results from three independent experiments. Values are relative to wild type, bars represent the mean of samples run in triplicate; all values are significant with P<0.05. EXPRESSION / LABELING:
|
|
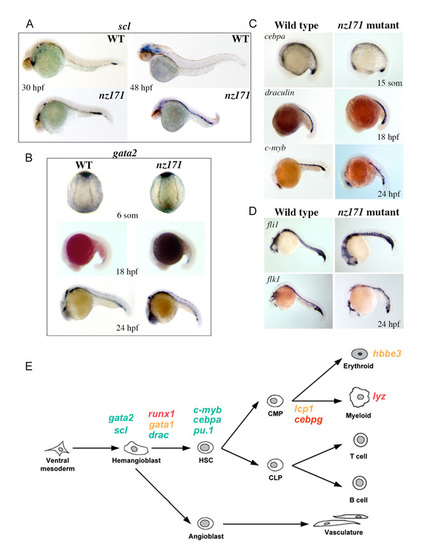
Most hematopoietic and vascular markers assessed are expressed normally in nz171 mutants. (A-D) Expression of hematopoietic markers in whole-mount embryos as detected by in situ hybridization. Anterior to the left (except B, top panels: posterior view). (A) scl expression in wild-type and nz171 mutant embryos, as indicated on the panels. At 30 h.p.f., expression levels of scl in nz171 embryos are similar to those in wild type, and at 48 h.p.f. scl expression is still observed in the ICM region, suggesting that it has not been downregulated as observed in wild-type embryos. (B) gata2 expression in 6-somite, 18 h.p.f. and 24 h.p.f. wild-type and nz171 embryos as indicated on the panels. gata2 transcript levels in nz171 mutants are similar to wild type levels. (C) Expression of various hematopoietic genes in wild-type and nz171 mutant embryos, as indicated on the panels. The presence of myeloid precursors in the ALM of nz171 embryos is indicated by the normal expression of cebpa and c-myb in this area. (D) Vascular markers fli1 and flk1 are expressed relatively normally in nz171 mutants at 24 h.p.f. (E) Schematic illustrating the expression status of hematopoietic genes in nz171 mutants according to when they are expressed developmentally. Green, gene expression not affected; orange, reduced; red, completely absent. EXPRESSION / LABELING:
|
|
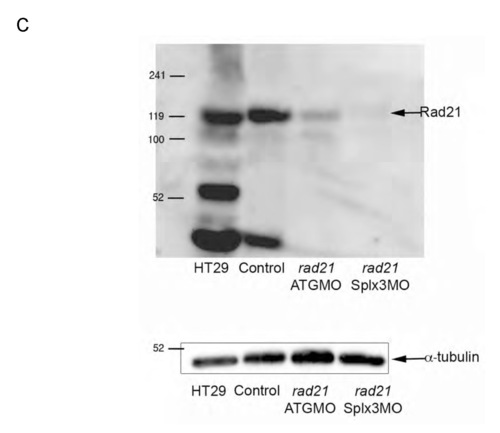
cDNA and protein sequence of Rad21nz171 and previously characterized alleles. The rad21 transcript contains 2615 nt and encodes a protein of 643 residues (NCBI). There are two chromosome 16- associated rad21 sequences available at NCBI: NM_199595/BC045311 and AY423040/Ensembl Transcript ENSDART00000005927. An alignment of proteins encoded by these two transcripts revealed a polymorphism at position 592, where glycine is substituted for serine in NM_199595/BC045311. (A) Annotated sequence of the cDNA synthesized from rad21nz171 homozygotes. The sequence is 2428 bp and incorporates the entire open reading frame. We designed antisense morpholino oligonucleotides directed against the rad21 transcript: two targeting the start and 5′ regions, and one targeting the 5′ donor of exon 3. Morpholino binding sites are indicated by highlighting: UTR_MO, yellow; ATG_MO, green; SPLX3_MO, pink. The start codon is boxed and the stop codon is indicated in bold. The G->T mutation in the rad21nz171 allele is boxed and highlighted in red. (B) Comparison of the protein sequence of two Rad21 sequences deposited in GenBank, NM_199595 and AY423040, with the Rad21nz171 protein sequence (Rad21 17.1). The stop codon at position 277 created by the mutation is indicated by an asterisk. The 276 amino acid truncated protein resulting from this mutation is predicted to be non-functional, as the C-terminal winged helix domain important for contacting the Smc1 head (Nasmyth and Haering, 2005) is deleted. Rad21nz171 and AY423040 both have serine at position 592, whereas NM_199595 has glycine. The hydrophilic nature of this region is conserved. (C) Top, immunoblot showing lack of Rad21 protein in rad21 morphants, indicating correct targeting of rad21 mRNA by morpholinos. The Rad21 antibody (AB3233, Chemicon) is directed to the C-terminus of the protein and is not expected to recognize truncated products. HT29 is a human colon carcinoma cell line; 5 μg total protein was loaded as a positive control for the anti-Rad21 antibody. ‘Control’ refers to non-injected wild-type embryos. Embryos were harvested at 48 hpf, and 20 μg total protein was loaded per lane. Bottom, anti-α-tubulin immunoblot as a loading control. EXPRESSION / LABELING:
|
|
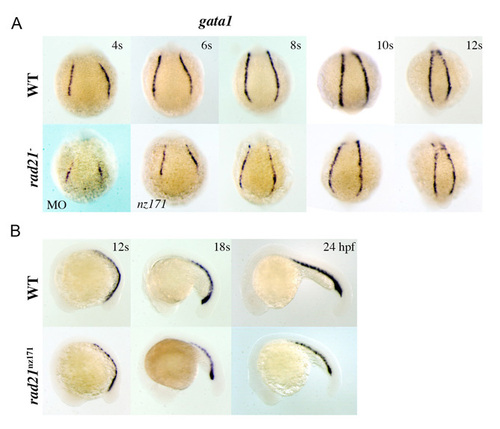
gata1 expression levels are reduced in rad21 morphants and mutants during early embryogenesis. Whole-mount embryos stained for gata1 expression. Upper panels are wild type; lower panels are rad21ATGMO morphants (MO) or rad21nz171 homozygotes as indicated. (A) Posterior views of embryos, stages as indicated. Onset of expression of gata1 is slightly delayed in rad21 morphants, as evidenced by gata1 expression in the 6s morphant appearing similar to its expression in the 4s wild-type embryo. All rad21 morpholino-injected embryos (rad21ATGMO; 0.5 pmol) at 4s (n=29/29) and 6s (n=20/20) had reduced/delayed expression of gata1. On close inspection of rad21nz171 mutants, gata1 expression appears to be ‘patchy’ in intensity within the PLM stripes. (B) Lateral views, anterior to the left, stages as indicated. gata1 expression is noticeably reduced in the PLM/ICM of rad21nz171 mutants. EXPRESSION / LABELING:
|
|
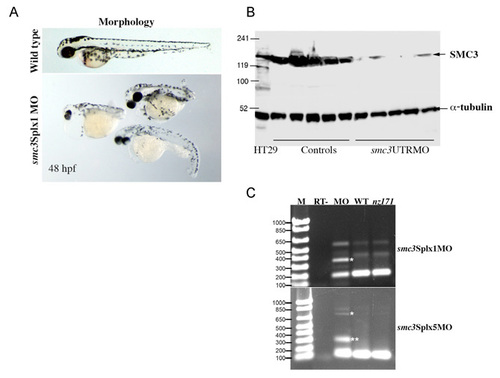
Targeted knockdown of the smc3 gene. Antisense morpholinos were designed to target the 5′ UTR (smc3UTRMO), the ATG start codon (smc3ATGMO), the splice site of exon 1, 3′ donor (smc3Splx1MO) and the splice site of exon 5, 3′ donor (smc3Splx5MO) of the smc3 mRNA. Ensembl locus ENSDARG00000019000 and sequence BC044408 were used to inform morpholino oligonucleotide design. Embryos were able to develop to 48 h.p.f. when given a 0.25-0.5 pmol dose of morpholino targeting smc3. Higher doses of 2-3 pmol caused arrest in early blastula stages. Total knockdown of Smc3 function should prevent cell division altogether. Therefore, post-blastula embryos are considered to be smc3 hypomorphs rather than smc3 nulls. (A) Representative gross morphology of Smc3 morphants at 48 hpf generated by a 0.5 pmol dose of smc3Splx1MO, compared with a wild-type embryo of the same stage (top). All morpholinos targeting smc3 generated a similar phenotype. (B) Representative anti-Smc3 immunoblot of protein obtained from embryos injected with 3 pmol smc3UTRMO. Individual embryos that had arrested at the early blastula stage were lysed and the entire sample was used for immunoblotting along with stage-matched controls treated in the same manner. The blot was cut in two and treated with anti-Smc3 antibody (Chemicon, 1:1000) or anti-α-tubulin (Sigma, 1:2000) to provide a loading control. 5 μg of protein from HT29 colon carcinoma cells was used as a positive control for the antibodies. (C) Splice-site morpholinos smc3Splx1MO and smc3Splx5MO are effective in blocking splicing of the smc3 mRNA, and show partial activity in smc3 hypomorph morphants. Agarose gel electrophoresis of PCR products obtained by RT-PCR of cDNA obtained from 24-hpf embryos. M, DNA size marker with number of base pairs indicated at left. RT-, control, morphant embryos processed without reverse transcriptase; MO, embryos injected with 2 pmol of smc3 morpholino (identity indicated at side); WT, wild-type embryos; nz171, rad21nz171 mutant embryos. White asterisks indicate bands of expected size should the intron involved at each splice junction remain unspliced. Double asterisk, band of unpredicted size. Primer sequences for RT-PCR: smc3Splx1 forward, 5′-CTGAGGAGTGTTTTGTGC-3′ and reverse, 5′-GATGACATTGTGTTTTGAGC-3′; smc3Splx5 forward, 5′-CGAGTCATTTCAGCCTTCG-3′ and reverse, 5′-TTACTGCGGGAGAATCCAG-3′. |

Unillustrated author statements EXPRESSION / LABELING:
PHENOTYPE:
|