- Title
-
Differentiation of cerebellar cell identities in absence of Fgf signalling in zebrafish Otx morphants
- Authors
- Foucher, I., Mione, M., Simeone, A., Acampora, D., Bally-Cuif, L., and Houart, C.
- Source
- Full text @ Development
|
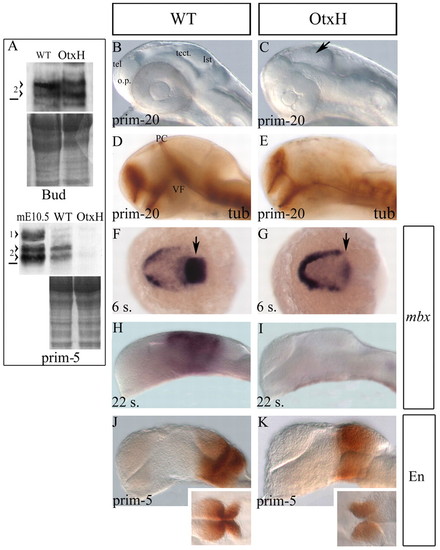
Absence of mesencephalon in OtxH embryos. (A) Western blot analysis on wild-type or OtxH zebrafish embryo extracts from end of gastrulation stage (upper panel: bud stage) or late somitogenesis stage (lower panel: prim-5). Proteins staining is shown underneath each to control protein levels between wild-type and OtxH embryos. 1>, mouse Otx1protein; 2>, doublet of mouse or zebrafish Otx2 proteins. The horizontal black line represents the 36.4 kDa marker. In OtxH embryos, Otx2 expression is not visibly affected at the end of epiboly but is lost by the end of somitogenesis. (B-E) Lateral views, anterior towards the left, of prim-20 (33 hpf) live brains (B,C) or fixed after acetylated tubulin staining (D,E) from wild-type (B,D) or OtxH (C,E) embryos. The isthmic constriction (arrow in C), the ventral flexure (VF) and the posterior commissure (PC, normally forming at the boundary between forebrain and midbrain) are all absent. tel, telencephalon; o.p., olfactory placode; tect., tectum; ist, isthmus. Dorsal (F,G) and lateral (H-K) views, anterior towards the left, of wild-type (F,H,J) or OtxH (G,I,K) brains. Expression of mbx is almost undetectable in the CNS at the six-somite stage in OtxH embryos (arrow in G) compared with wild type (arrow in F) and is lost by the 22-somite stage (I). Expression of Engrailed is detectable in both wild type (J) and OtxH (K) at prim-5 stage. Insets are dorsal views of the isthmic area of the brains shown in J and K. EXPRESSION / LABELING:
|
|
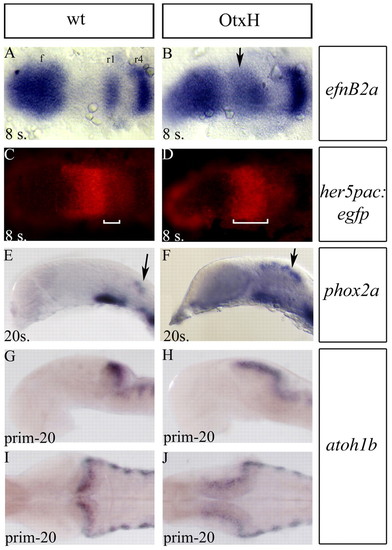
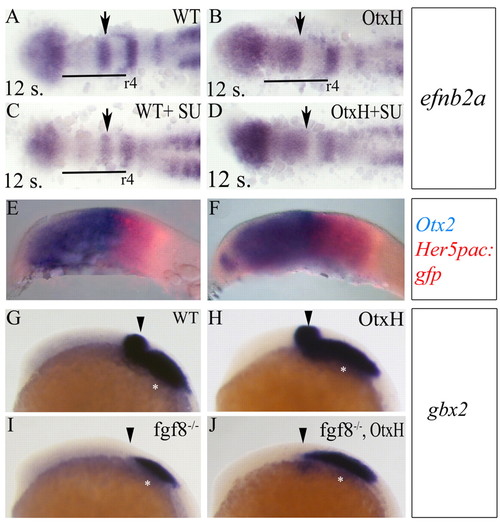
Mesencephalon to rhombomere1 transformation in OtxH embryos. (A-D) Dorsal views (anterior towards the left) of eight-somite embryos after double detection of efnb2a (blue in A,B) expressed in forebrain (f), rhombomeres 1 (r1), 4 (r4) and 7 (r7) and GFP (red in C,D) in wild type; her5pac:egfp (A,C) and OtxH; her5pac:egfp (B,D). In OtxH, the mesencephalon is transformed into an expanded r1 territory (white brackets in C and D indicate the anteroposterior extent of efnb2a). (E-J) Lateral (E-H) and dorsal (I,J) views, anterior towards the left, of wild-type (E,G,I) and OtxH (F,H,J) brains. Locus coeruleus cells expressing phox2a (arrows in E,F) and rhombic lip cells expressing atoh1b (G-J), which is known to arise from the r1 territory, are expanded in OtxH embryos (F,H,J). I and J are dorsal views of embryos in G and H. EXPRESSION / LABELING:
|
|
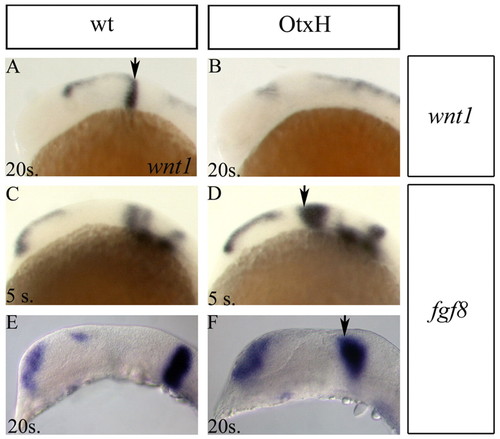
Isthmic organiser defects. Lateral views of wild-type (A,C,E) and OtxH (B,D,F) brains. (A,B) Most of wnt1 expression is lost in OtxH embryos; weak remaining expression is often observed anterior to the expanded r1. (C-F) fgf8 expression is expanded in the dorsal mes/met territory of OtxH embryos (arrows) at the 5-(D) and 20-somite stages (F). Arrows indicate the isthmic expression, expanded in the morphants. EXPRESSION / LABELING:
|
|
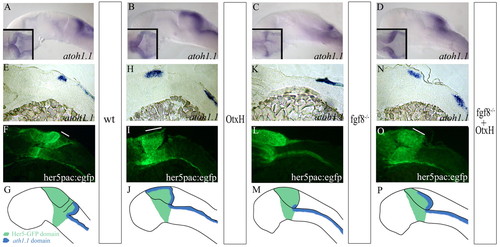
Formation of the granule cell population in fgf8-/-; OtxH embryos. atoh1a expression in wild-type (A), OtxH (B), fgf8-/- (C) and fgf8-/-; OtxH (D) whole-mount embryos. MyoD expression in the somite was concomitantly detected to confirm fgf8-/-genotype. (E-O) Para-saggital 10 μm sections, anterior towards the left, of wild-type her5pac:egfp (E,F), OtxH (H,I), fgf8-/- (K,L) and fgf8-/-; OtxH (N,O) brains, showing co-localisation of atoh1a (E,H,K,N) and GFP expression (F,I,L,O). Granule cells absent in fgf8-/- are rescued in fgf8-/-; OtxH and are GFP positive (white lines on fluorescent pictures represent the length of the I>atoh1a domain in the upper rhombic lip obtained from bright field pictures). (G,J,M,P) Cartoons summarising GFP (in green) and atoh1a (in blue) expressions in wild-type (G), OtxH (J), fgf8-/- (M) and fgf8-/-; OtxH (P) embryos carrying the transgene her5pac:egfp. EXPRESSION / LABELING:
|
|
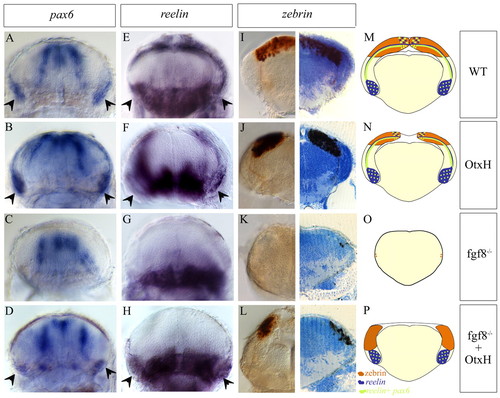
Cerebellum formation in fgf8-/-; OtxH embryos at 5 days post-fertilization. Transverse sections of cerebellar areas from wild-type (A,E,I), OtxH (B,F,J), fgf8-/- (C,G,K) and fgf8-/-; OtxH (D,H,L) embryos showing expression of pax6 and reelin in granule cells (arrowheads in A-D and E-H, respectively) and zebrin in Purkinje cells (I-L). A total number of 10, 4 and 12 fgf8-/-; OtxH embryos were analysed for pax6, reelin and zebrin staining, respectively. All the fgf8-/-; OtxH embryos show the rescued phenotype. The count of zebrin-positive cells on vibratome 15 μm sections shows that a very small number of Purkinje cells are sometimes present [fgf8-/- embryos have 2.4±2.4 positive cells (n=4 embryos counted) whereas fgf8-/-; OtxH embryos have 15±5 Purkinje cells per section (n=4)]. Cartoons represent transverse section through the cerebellum in wild type (M), OtxH (N), fgf8-/- (O) and fgf8-/-; OtxH P). Zebrin-positive cells are brow; reelin and reelin+ pax6 granule cells are blue and green, respectively. Only a subset of reelin cells is positive for pax6. Granule cells are always missing in fgf8-/- (C,G). Ventral reelin staining corresponds to glial cells of the reticular formation, whereas proliferative regions of the midline at the level of the IVth ventricule and two lateral stripes of radial glia are pax6 positive. EXPRESSION / LABELING:
|
|
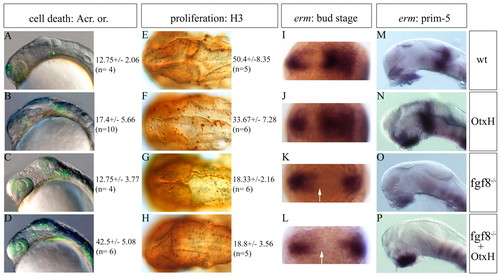
fgf8-/-; OtxH embryos have a total lack of Fgf activity and show both a proliferation decrease and a cell death increase. (A-D) Lateral view of 20-somite stage live embryos stained with Acridine Orange and cell quantification (n, numbers of quantified embryos). No change in cell death is observed in fgf8-/- mutant embryos (compare A with C), while OtxH shows an increase in dying cells (B), which is enhanced in fgf8-/-; OtxH (D). These differences are more drastic at prim-22 stage (wild type: 18±4, n=3; OtxH 26±2, n=3; fgf8-/-: 12±2, n=3; fgf8-/-; OtxH: 44.33±6.03, n=3). (E-H) Dorsal views of prim-22 stage posterior brain immunostained with the H3 reveal a decrease in proliferation rate in fgf8-/- mutant (G), while lack of Otx in this context does not worsen the phenotype (H), see also quantifications. (I-P) erm expression pattern in the mid/hindbrain region of the genetic context studied at bud stage (dorsal view, I-L) and at prim-5 stage (lateral view, M-P). This Fgf transcriptional target is never induced in the presumptive mes/met (arrow) in fgf8-/- (K,O) or fgf8-/-; OtxH (L,P) embryos. |
|
Early r1 rescue in fgf8-/-; OtxH embryos is independent of gbx2. Dorsal views, anterior towards the left showing efnb2a expression in wild-type (A, n=5/5), OtxH (B, n=8/9), wild type+SU5402 (C, n=12/15) or OtxH+SU5402 (D, n=18/23) embryos. The anteroposterior extent of r1 territory (arrows) is the same in OtxH with or without SU5402. Conversely, SU5402 treatment affects r5-7 development as previously reported (Maves et al., 2002). (E,F) Lateral views, anterior towards the left, of wild-type (E) and fgf8-/- (F) 8-somite stage embryos, showing double expression of otx2 (blue) and her5pac:egfp (red). (G-J) Lateral views, anterior towards the left, showing gbx2 expression in wild type (G), OtxH (H), fgf8-/- (I) or fgf8-/-; OtxH (J). gbx2 CNS expression is lost in fgf8-/- (arrows) and is not rescued in fgf8-/-; OtxH (out of 21 fgf8-/-; OtxH embryos, only five show a very faint rescue of Gbx2 expression in the very ventral metencephalic area). Asterisks indicate gbx2 expression in the epibranchial placode precursors. EXPRESSION / LABELING:
|
|
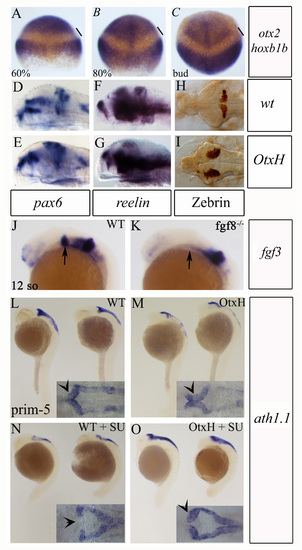
Invariability in position of the Otx2 posterior expression boundary. Double detection of otx2 and hoxb1b transcripts in wild-type embryos at 60% (A), 80% (B) and bud (C) stages. hoxb1b anterior limit defines the r3/r4 boundary. The gap between the two expression domains remains constant, arguing against an anterior shift in otx2 expression during late gastrulation. (D-I) Rostral expansion of cerebellum in OtH embryos. Lateral (D-G) or dorsal (H,I) views of cerebellar areas from wild-type (D,F,H) and OtxH (E,G,I) embryos, showing expansion of pax6 (D,E) and reelin (F,G) expression in granule cells, and zebrin (H,I) in Purkinje cells in OtxH embryos. (J-O) Rescue of granular cells in OtxH embryos lacking Fgf signal. (J-K) Lateral views, anterior towards the left, of 12-somite stage brains showing fgf3 expression, which is never detectable in the isthmus of fgf8–/– mutant embryos (arrows indicate the presumptive isthmic area). (L-O) Lateral views of wild-type (L), OtxH (M), wild-type treated with SU5402 (N) and OtxH treated with SU5402 (O) whole-mount embryos, showing atoh1a expression. Granule cell precursors in the upper rhombic lip in wild type (arrow on dorsal view in inset L) are lost after inhibition of Fgf signal (arrow on dorsal view in inset N). The residual expression in lateral and lower rhombic lip is not giving rise to granule precursors. The upper rhombic lip population increases in OtxH (arrow on dorsal view in inset M), and is kept in OtxH+SU5402 (arrow on dorsal view in inset O). |
|
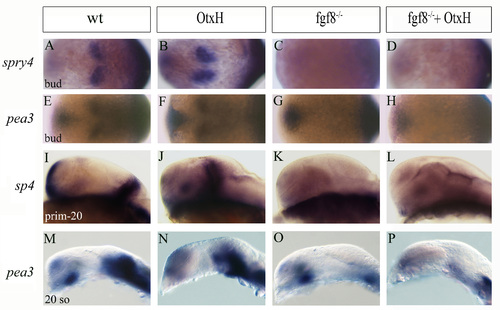
Absence of Fgf targets in fgf8–/– and fgf8–/–; OtxH embryos.(A-P) sprouty 4 (A-D,I-L) and pea3 (E-H,M-P) expression pattern in the mid/hindbrain region of the genetic context studied at bud (A-H), 20-somite (M-P) and prim-5 stages (I-L). These Fgf transcriptional targets are never induced in fgf8–/– or fgf8–/–; OtxH embryos (see D,H,L,P) |
|
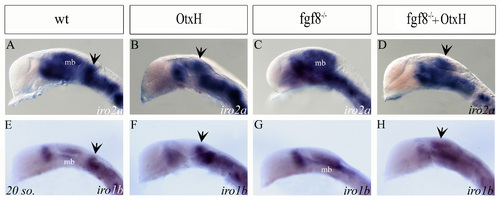
Expression profile of iro2a and iro1b. All prim-5 brains are lateral view, anterior to the left. Expression of iro2a (A-D) and iro1b (E-H) in wild type (A,E), OtxH (B,F), fgf8–/– (C,G) and fgf8–/–; OtxH (D,H) embryos. Both genes are expressed in diencephalon subdomains, midbrain (mb), r1 and hindbrain in wild type. Diencephalic and midbrain domains are, respectively, reduced or lacking in OtxH embryos. A fainter midbrain domain is expanded in fgf8–/– embryos. Prospective cerebellar territories are iros-positive in wild type, OtxH and fgf8–/–; OtxH embryos (arrows), while OtxH embryos might have a general slight decrease in mid/hinbrain staining. EXPRESSION / LABELING:
|

Unillustrated author statements EXPRESSION / LABELING:
|