- Title
-
sox4b is a key player of pancreatic alpha cell differentiation in zebrafish
- Authors
- Mavropoulos, A., Devos, N., Biemar, F., Zecchin, E., Argenton, F., Edlund, H., Motte, P., Martial, J.A., and Peers, B.
- Source
- Full text @ Dev. Biol.
|
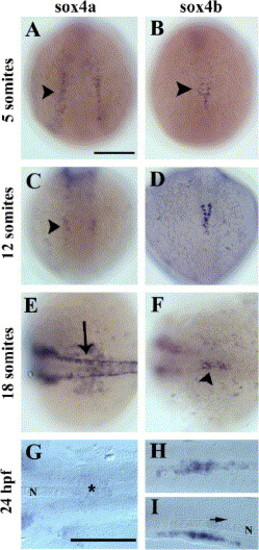
Expression of sox4a and sox4b genes during zebrafish development. Whole-mount in situ hybridization with sox4a- (left panels) and sox4b- (right panels) digoxigenin-labeled anti-sense probes at various stages of zebrafish development (from the 5-somite stage to 24 hpf). (A–D) Dorsal views, anterior to the top. (E, F) Dorsal views, anterior to the left. (G–I) dorsal views (G, H) and lateral view (I) with anterior to the left of yolk-free embryos. A scale bar = 200 μm, G scale bar = 200 μm in G, H, and I. Arrowhead in panel B: mid-trunk endoderm. Arrow in panel E: dorsal neural tube; arrow in panel I: floor plate. The notochord (N) is indicated in panels G and I. As soon as the 5-somite stage (B), sox4b starts to be expressed in some scattered cells of the mid-trunk endoderm (arrowhead). As somitogenesis goes on, the expression persists in this region (D, F arrowhead) until 24 hpf (H, I). By contrast, sox4a is expressed in various tissues such as the lateral plate mesoderm (A, arrowhead), the hindbrain (C, arrowhead), the neural crest (E, arrow) but expression in the pancreatic region is hardly above the detection limit (G, star). EXPRESSION / LABELING:
|
|
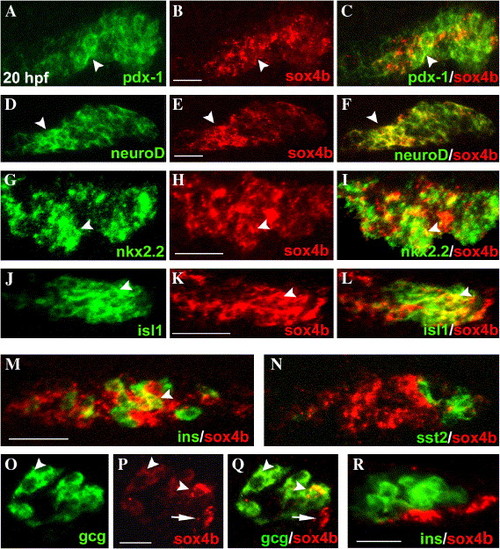
Expression of sox4b in pancreas observed by FISH. (A–L) Expression of either pdx-1, neuroD, nkx2.2, isl1 (left column), or sox4b (central column) in 20 hpf embryos. The right column presents the overlay of the respective left and middle panels. (M, N) Overlay of sox4b- and either insulin- (M) or somatostatin2 (N) expression in 20 hpf embryos. (O, P) Expression of glucagon (O) and sox4b (P) in 36 hpf embryos. (Q) Overlay of panels O and P. (Q) Overlay of sox4b- and insulin expression in 26 hpf embryos. All the panels are optical sections of ventrally viewed yolk-free embryos (A–Q) and laterally viewed yolk-free embryo (R), with anterior to the left. sox4b was detected with a digoxigenin-labeled anti-sense probe and a tyramide-Cy3 substrate and the other markers with DNP-labeled anti-sense probes and tyramide-FITC substrate (see Materials and methods). A–N scale bars = 50 μm; O–Q scale bars = 25 μm. The arrowheads point to cells where sox4b is co-expressed with another marker, arrows in panels P and Q point to cells expressing only sox4b. EXPRESSION / LABELING:
|
|
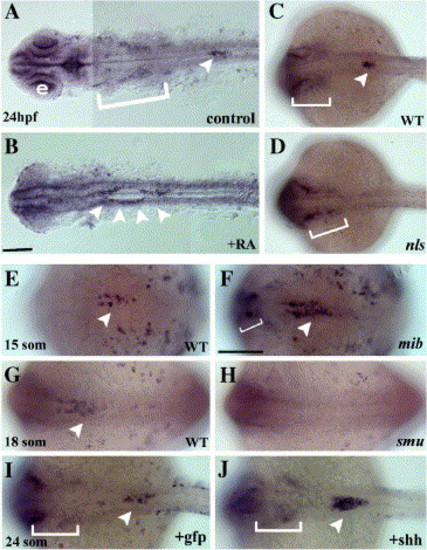
Expression of sox4b in response to affected retinoic acid, Hedgehog and Delta/Notch signaling. Dorsal views of whole-mount in situ hybridization for sox4b expression. Anterior to the left; B and F scale bars = 100 μm in A–D panels and in panels E–G, respectively. Arrowheads and brackets indicate sox4b expression in pancreas and in neural crest, respectively. Panels A and B present yolk-free embryos. e: eye. Exogenous RA leads to an anterior shift of sox4b expression in pancreas, as described for pancreatic markers (A: control embryo, B: RA-treated embryos). In the nls mutant, defective in RA synthesis, sox4b expression is absent from the pancreatic region (C: wild-type sibling embryo, D: nls mutant embryo). Up-regulation of sox4b is observed in the mib mutant embryos (E: wild-type sibling, F: mib mutant embryo), defective in the Delta-Notch signaling pathway. Alterations of the Hedgehog signaling affect also sox4b expression in pancreas (G–J). In the smu mutant embryos, in which the endocrine part of pancreas fails to develop, sox4b is absent (G: wild-type sibling, H: smu mutant embryo). In addition, injection of shh mRNA induces the over-expression of sox4b (I: GFP mRNA-injected embryo, J: shh mRNA-injected embryo), as previously shown for endocrine pancreatic markers. EXPRESSION / LABELING:
|
|
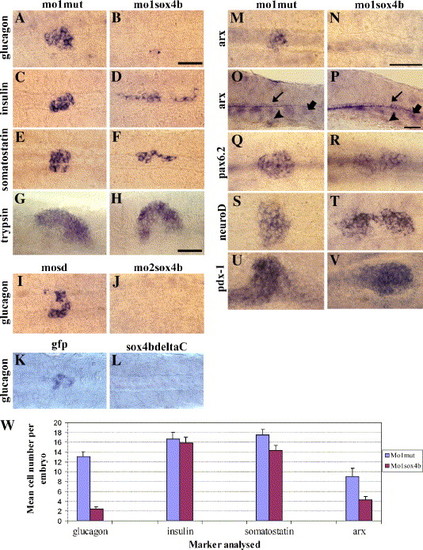
sox4b is required for the differentiation of α cells. (A–V) Expression of various pancreatic markers in wild-type control embryos (first and third columns) or sox4b knock-down embryos (second and fourth columns), analyzed at 30 hpf (except trypsin that was analyzed at 3 dpf). Compared to the controls (A and I), glucagon expression dramatically decreases in embryos injected with either mo1sox4b or mo2sox4b (B and J). The injection of the mRNA encoding Sox4deltaC leads to the same phenotype (K and L) while the injection of gfp mRNA has not any effect. By contrast, the expression of insulin (C, D), somatostatin (E, F) is not much affected although the cells are scattered. In addition to glucagon decrease, sox4b knock-down causes a similar phenotype on arx expression specifically in the pancreas (panels M and N and arrowhead in O and P), without any effect on the floor plate nor the somites (thin and thick arrows in panels O and P, respectively). By contrast, the levels of expression of pax6.2 (Q, R), neuroD (S, T), pdx-1 (U, V), and trypsin (G, H) are not significantly affected. All the panels present yolk-free embryos. Except panels O and P that are lateral views, all the panels are dorsal views with the anterior to the left. B, N, and P scale bars = 50 μm; H scale bar = 100 μm. In panels O and P, arrowhead: dorsal bud, thin arrow: floor plate, thick arrow: somites. W: quantitative analysis of endocrine markers in mo1mis or mo1sox4b morphant embryos at 30 hpf. The knock-down of sox4b leads, on average, to the loss of 82% and 55% of glucagon and arx cells. The average number of cells of the other markers analyzed is poorly affected. Y bars correspond to the standard error. See Table 1 for embryo numbers. EXPRESSION / LABELING:
PHENOTYPE:
|
Reprinted from Developmental Biology, 285(1), Mavropoulos, A., Devos, N., Biemar, F., Zecchin, E., Argenton, F., Edlund, H., Motte, P., Martial, J.A., and Peers, B., sox4b is a key player of pancreatic alpha cell differentiation in zebrafish, 211-223, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.