- Title
-
Dual roles of zygotic and maternal Scribble1 in neural migration and convergent extension movements in zebrafish embryos
- Authors
- Wada, H., Iwasaki, M., Sato, T., Masai, I., Nishiwaki, Y., Tanaka, H., Sato, A., Nojima, Y., and Okamoto, H.
- Source
- Full text @ Development
|
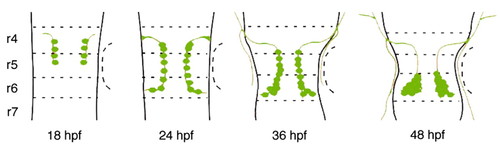
Schematic drawing of migration of the nVII motor neurons in zebrafish. Dorsal views of the zebrafish hindbrain at each developmental stage. The nVII motor neurons and their axons are shown in green. Broken lines indicate rhombomeric boundaries. See text for details. |
|
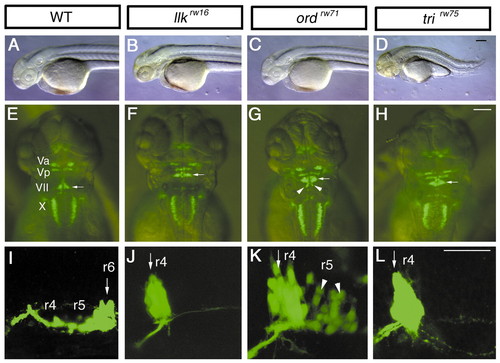
Isolation of mutants with disrupted migration of the nVII motor neurons. Morphology and Isl1-GFP expression in the wild-type (A,E,I), llkrw16 (B,F,J), ord rw71 (C,G,K), and tri rw75 homozygous embryos (D,H,L) at 2 dpf (A-H) and 30 hpf (I-L). In the wild-type embryos, the nVII motor neurons are located in r6 (E,I, arrows). In contrast, in the llk rw16 and tri rw75 embryos the nVII motor neurons are located in r4 (F,J,H,L, arrows). In the ord rw71 embryos, the nVII motor neurons are located in r4 and r5 (G,K, arrowheads indicate cells migrating into r5). The cells that migrated into r5 became detached from the surface of hindbrain, and were scattered inside the hindbrain. The tri rw75 embryos show severe defects in the extension of the trunk region (D). The llk and ord embryos did not show any morphological abnormalities, in contrast to the tri embryos (B,C). (A-D, I-L) Lateral views; anterior is to the left, (E-H) dorsal views; anterior is to the top. Scale bars: 50 µm.
|
|
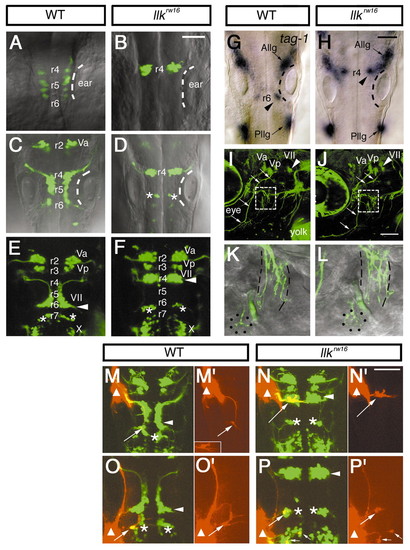
Migration of the nVII motor neurons is specifically impaired in llk embryos. A-F, Isl1-GFP expression in the wild-type (A,C,E) and llk rw16 homozygous embryos (B,D,F) at 18 hpf (A,B), 24 hpf (C,D), and 48 hpf (E,F). In the wild-type siblings, the nVII motor neurons arise in r4, migrate caudally through r5 into r6 and form the nucleus in r6 (E, arrowhead). In contrast, in the llkrw16 embryos the GFP-expressing cells that arise in r4 fail to migrate and form an ectopic nucleus in r4 (F, arrowhead). Asterisks (D,E,F) indicate r6-derived putative OLe neurons, which migrate into r7 in the wild-type embryos. These neurons also fail to migrate in the mutant embryos and remain in r6 (D,F). (G,H) tag-1 mRNA expression in the wild-type (G) and llk rw16 (H) embryos at 24 hpf. tag-1-positive cells are located in r4 in the llkrw16 embryo (H, arrowhead). Dorsal views. The position of the ears are indicated by the broken lines. Va, Vp, anterior and posterior trigeminal nuclei, respectively; VII, facial nucleus; X, vagus nucleus; Allg, Pllg, anterior and posterior lateral line ganglion, respectively. (I-L) Isl1-GFP expression in the wild-type (I,K) and llk rw16 (J,L) embryos at 5 dpf. Arrowheads indicate the facial motor nucleus. The trajectories of facial motor axons are normal in the llk rw16 embryo (arrows). The axons reach the target organs (K,L, higher magnifications of the boxed regions in I and J). Lateral line organs and the cranial muscles are indicated by dots and broken lines, respectively. Lateral views; anterior is to the left. (M-N) The putative OLe neurons (arrows) projecting to the lateral lines are retrogradely labeled (red) in the wild-type (M,O) and llk rw16 (N,P) embryos. DiI was applied to the anterior (M,N) or posterior lateral line ganglion (O,P). Sites of DiI application are indicated by triangles. Small arrows indicate the vagal (X) motor neurons, which were labeled with DiI that diffused from the application site (P). Single-channel images of the labeled neurons are shown in M′, N′, O′ and P′. These neurons extend dendrites to the contralateral side of the brain. Inset in M′ shows another example in which dendritic processes were clearly labeled. Asterisks indicate r6-derived r7-located neurons, which fail to migrate and remain in r6 in the llk embryos. Arrowheads indicate the facial motor nuclei. Dorsal views. Scale bar: 50 µm.
EXPRESSION / LABELING:
PHENOTYPE:
|
|
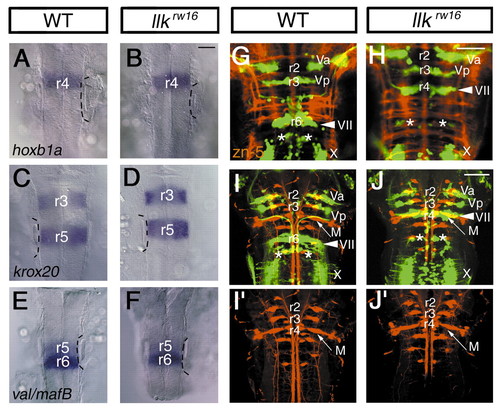
Rhombomeric patterning and differentiation of the neurons are unaffected in the llk embryo. (A,B) hoxb1a; (C,D) krox20; (E,F) valentino/mafB mRNA expression in the wild-type (A,C,E) and llk rw16 (B,D,F) embryos at 20 hpf. Expression patterns of these genes are unaffected in the llk rw16 embryo. Dorsal views. The position of the ears are indicated with broken lines. (G,H) Commissural axons are labeled with zn-5 antibody (red) in the wild-type (G) and llk rw16 (H) embryos at 36 hpf. (I,J) Reticulospinal neurons are retrogradely labeled (red) in the wild-type (I) and llk rw16 (J) embryos at 5 dpf. Single-channel images of the labeled neurons are shown in separate panels (I′,J′). M, Mauthner′s cell. Asterisks indicate r6-derived putative OLe neurons. Arrowheads indicate the facial motor nuclei. Dorsal views. Scale bar: 50 µm. EXPRESSION / LABELING:
|
|
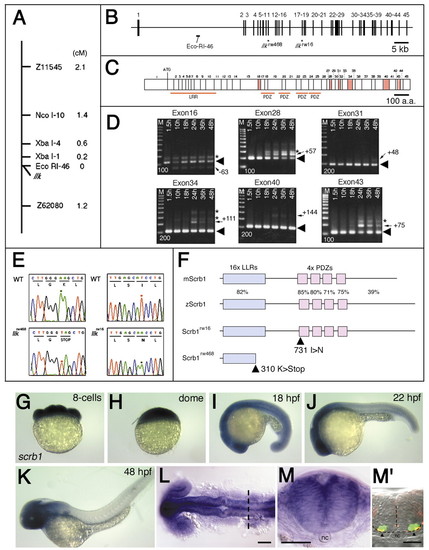
Identification of the llk gene. (A) Genetic map for the llk locus. The llk locus mapped to linkage group (LG) 7. The RDA marker, EcoRI-46 is located at 0 cM map-distance from the llk locus (0 per 2054 meioses). (B) Genomic structure of the zebrafish scrb1. The scrb1 gene is encoded by 45 exons spanning 120 kb in the genome. EcoRI-46 is located in the first intron of the gene. Each of the two mutant alleles, lklrw16 and llkrw468, carries a nucleotide substitution in exons 11 and 17, respectively. (C) Schematic drawing for the putative cDNA encoding 45 exons shown in B. Six alternatively used exons, 16, 28, 31, 34, 40 and 43 are shown as red boxes. Exon numbers and first ATG site are indicated above. Regions encoding LRR domain and four PDZ domains are indicated below. (D) RT-PCR analyses were performed to identify the predominant gene product. Primers were designed in the flanking exons encompassing each exon of interest. Total RNA was extracted from 1.5, 10, 18, 24, 36 and 48-hpf embryos. Arrowheads in each panel indicate the predominant RT-PCR product expressed during migration of the nVII motor neurons at 18-24 hpf. The predominant RT-PCR products contain exon 16, but no other exons (exons 28, 31, 34 and 43). The lesser RT-PCR products contain exons 28, 31, 34 and 43, but not exon 16 (indicated by arrows). 100 bp-interval molecular markers (bp) are shown in each panel. (E) Sequence diagrams of the mutation sites for the llkrw16 and llkrw468 alleles compared to the wild-type allele. (F) Schematic drawings of the wild-type (zScrb1) and mutant Scrb1 proteins (Scrbrw16 and Scrbrw468). Percentage identity of the amino acid sequences (%) to the mouse Scrb (mScrb1) is shown for each domain. The allele llkrw16 carries a mis-sense amino acid substitution in the first PDZ domain, while llkrw468 carries a stop codon in the LRR domain. (G-M) Lateral views of wild-type embryos stained with the scrb1 RNA probe in the 8-cell stage (G), dome-stage (H), 18 hpf (I), 22 hpf (J) and 48 hpf (K) embryos. (L,M) scrb1 mRNA expression in the brain at 20 hpf (L, dorsal view) and (M) cross section at r5 (indicated by the broken line in L). M′ shows the cross section at r5 of the 24 hpf-Isl1-GFP embryo stained with anti-acetylated α-tubulin antibody. Arrowheads indicate medial longitudinal fascicles (MLF); nc indicates notochord. Scale bars: 50 µm. |
|
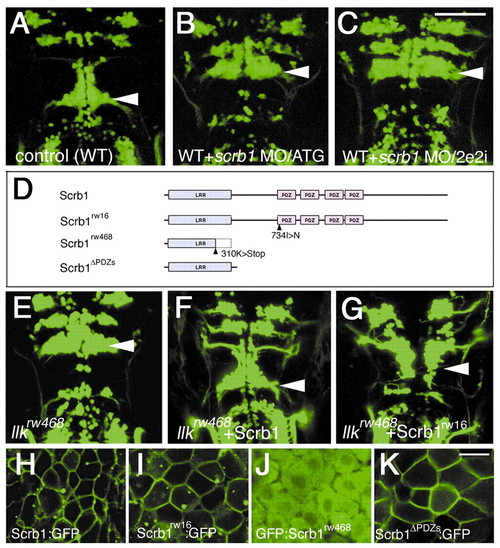
Loss-of-function and gain-of-function of scrb1 confirm that scrb1 is homologous to the llk gene. (A-C) Embryos injected with MO did not show any migration of the nVII motor neurons. MOs were designed to disrupt the translation (B, MO/ATG) or splicing (C, MO/2e2i) of the scrb1 mRNAs (compare with the wild-type embryo shown in A). Dorsal views, 2 dpf. (D-G) Structure-function analyses of Scrb1. Wild-type and mutated scrb1 mRNAs (schematically drawn in D) were injected into llkrw468 embryos. Injection of wild-type scrb1 mRNA restored migration of the nVII motor neurons (F, compare with control llkrw468 embryo shown in E). Injection of scrb1rw16 mRNA also restored the migration (G) although at lower frequency. (H-K) Subcellular localization of wild-type and mutated Scrb1 proteins. Scrb1, Scrb1rw16 and Scrb1<ΔPDZs are associated with plasma membranes (H,I,K). However, Scrb1rw468 failed to localize to membranes (J). A-C, E-G, dorsal views, 2 dpf. H-K, 10-12 hpf. Scale bars: 50 µm (A-G) and 20 µm (H-K).
|
|
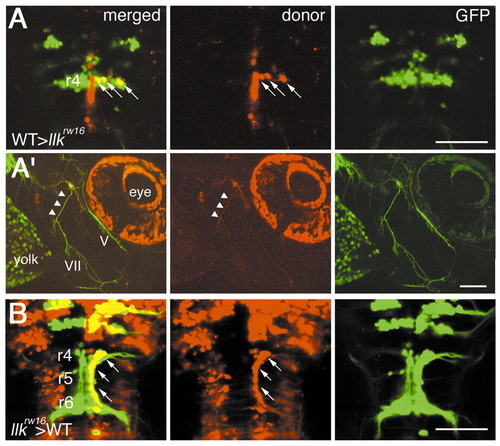
The llk gene is required for migration of the nVII motor neurons in a non cell-autonomous manner. Mosaic experiments were performed to determine the cell autonomy of the llk gene. A total of 8 wild-type embryo-derived nVII motor neurons (arrows) all failed to migrate caudally in 3 llk rw16 host embryos (A). (A′) The peripheral axons (arrowheads) of these cells were comprised in a part of the facial motor axons bundle. (B) In contrast, a total of 21 llk rw16 embryo-derived nVII motor neurons (arrows) migrated normally through r5 into r6 in 2 wild-type host embryos. (A,B) Dorsal views; (A′) lateral views, anterior is to the right, 2 dpf. Scale bar: 50 µm.
|
|
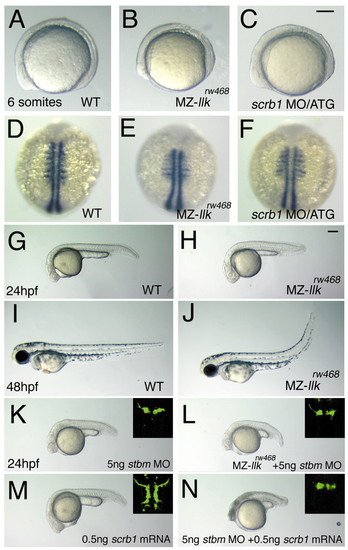
Maternal llk/scrb1 is required for convergent extension movements and genetically interacts with tri/stbm. (A-F) Maternal and zygotic (MZ-) llkrw468 embryos show slight convergent extension (CE) defects. Wild-type (A,D), MZ-llkrw468 (B,E) and scrb1 MO/ATG-injected (C,F) embryos were observed when alive (A-C) or labeled with myoD RNA probe(Weinberg et al., 1996) (D-F). In MZ-llkrw468 and scrb1 MO/ATG-injected embryos, the anterior-posterior axis was shorter and somatic mesoderm wider than wild-type embryos. (G-J) Morphology of embryos recovered in the later stages; only tail regions are deficient in MZ-llkrw468 embryos (H,J; compare with wild-type embryos shown in G,I). (K-N) llk/scrb1 genetically interacts with tri/stbm. (K) Wild-type embryos injected with stbm MO show slight CE defects. (L) MZ-llkrw468 embryos injected with stbm MO had slightly greater CE defects. (M) Wild-type embryos injected with scrb1 mRNA had slight CE defects. (N) Wild-type embryos co-injected with stbm MO and scrb1 mRNA showed severe CE defects. (K-L) Images of the nVII motor neurons in each embryo are shown in insets. Scale bars: 100 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
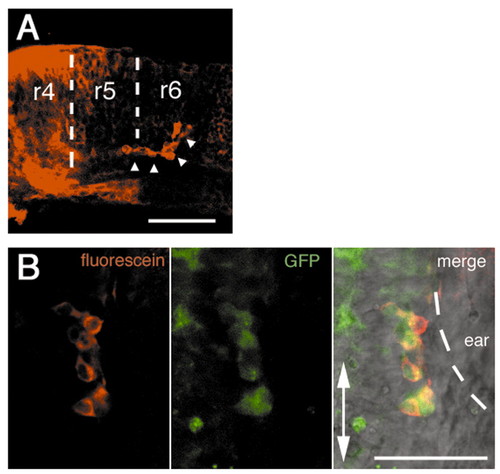
The nVII motor neurons migrate independently of the rest of the r4 tissues. The r4 region was labeled by uncaging the caged fluorescein-conjugated dextran and the cell movements were traced during development. (A) The nVII motor neurons that migrated into r5 and r6 (arrowheads) were the only population to come out of the labeled r4 tissue. Lateral view, anterior is to the left. (B) Double staining with anti-caged fluorescein and anti-GFP antibodies show that these r4-derived cells are the nVII motor neurons. Ventral views. Scale bars: 50 µm.
|
|
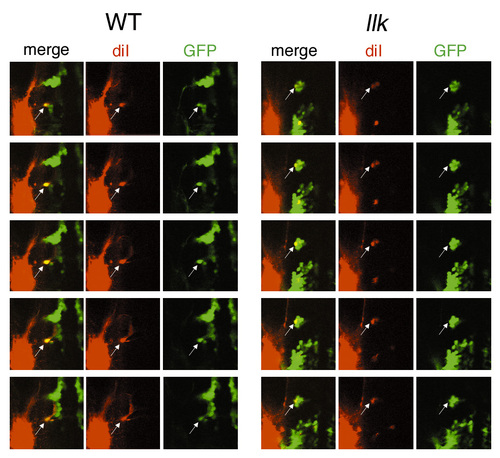
The putative OLe neurons projecting to the posterior lateral lines are retrogradely labeled in the wild-type and llk rw16 embryos. Original serial optical sections, from which stack images were synthesized for Fig. 3O,P. Images were obtained by confocal microscopy at 10 mm interval. Diameters of confocal pinholes to detect the red and green signals were identical (154 mm) to avoid mis-identification. In each focal plane, co-localization of DiI and GFP signals are shown (arrows). Thus, we conclude that some of the GFP-expressing neurons extend to the posterior lateral lines, and we referred to these cells as ?putative OLe neurons? in the text. |