- Title
-
Positional cloning of the young mutation identifies an essential role for the Brahma chromatin remodeling complex in mediating retinal cell differentiation
- Authors
- Gregg, R.G., Willer, G.B., Fadool, J.M., Dowling, J.E., and Link, B.A.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
yng is caused by a mutation in brg1. (a) Genetic and physical map. Linkage was initially discovered between simple sequence length polymorphism markers z1140 and z5623. Polymorphisms within ESTs mapped to this region on the T51 radiation hybrid panel and were then used to refine the interval. EST fb79a02 was found to contain no recombinants of 2,024 meioses, and BAC and PAC clones were used to build a contig around this locus. PACs 182p8 and 81j4 and BAC 38j4 were sequenced to reveal a high level of synteny with human chromosome 19 based on BLAST results. Positive rescue experiments using PAC 81j4 limited the search to either zebrafish ldlr or brg1. Vertical lines are exons determined by PCR of embryonic cDNA and based on GENSCAN predictions. (b) BAC/PAC rescue analysis. Transverse retinal sections showing the presence of TH (a late interplexiform cell marker, arrows) in a WT embryo injected with control buffer, a yng mutant injected with BAC 93m1 (which does not contain brg1 and is negative for TH expression), and a yng mutant injected with PAC 81j4 (which does contain brg1 and rescued TH expression). Arrows indicate retinal TH-positive neurons. TH is also normally expressed in the ventral forebrain of both yng and WT embryos (Right). (c) In situ mRNA analysis of brg1. Sixteen-cell stage embryos, a time before zygotic transcription begins, shows that brg1 transcript is provided maternally. Results with sense control and antisense riboprobes are shown. Whole-mount analysis at 48 hpf reveals strong anterior brg1 expression. Transverse retinal sections at 24, 48, and 60 hpf show robust brg1 expression during differentiation. (d) Sequence analysis and predicted protein structure. A C→ A transversion was identified in cDNA from yng homozygous mutants and later confirmed through genomic sequence analysis (red arrow). This base substitution changes a tyrosine codon to a stop codon very early with in the coding sequence of brg1 (red arrowhead). This truncation deletes all of the identified functional domains. |
|
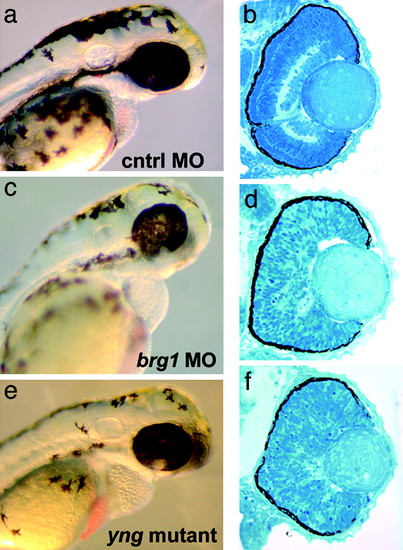
Morpholinos directed against brg1 phenocopy yng. (a and b) Standard control oligo. (c and d) Brg1MO1. (e and f) yng mutant, uninjected. Morphological inspection (a, c, and e) at 3 days postfertilization of Brg1MO1 and yng mutant embryos reveals similar defects in heart, ear, fin, and pigment cell development. Retinal histology (b, d, and f) shows that brg1 morpholinos also phenocopy the yng retinal lamination phenotype. The nonoverlapping oligo Brg1MO2 showed equivalent results to Brg1MO1. PHENOTYPE:
|
|
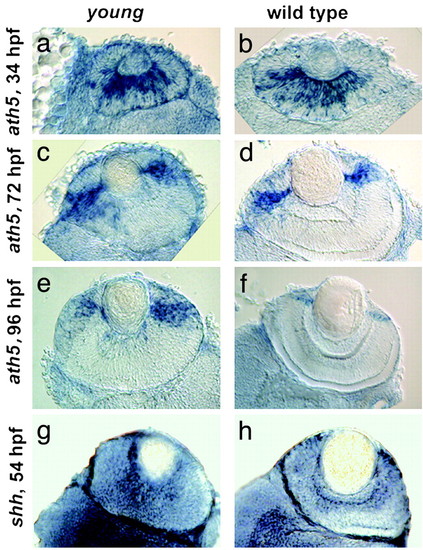
Expression of ath5 and shh is normal in yng retinas. In situ mRNA hybridizations of ath5 at 34, 72, and 96 hpf (a–f) and shh at 54 hpf (g and h) show similar patterns in yng (a, c, e, and g) and WT (b, d, f, and h) retinas. For each, whole-mount in situ hybridizations were performed and then transverse retinal sections were obtained. |
|
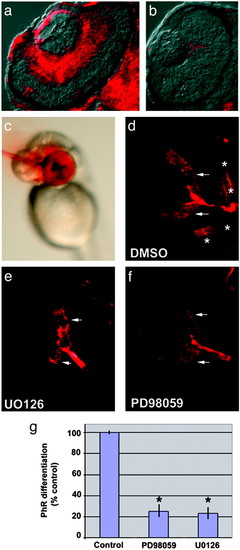
Disruption of MAP kinase activity blocks retinal differentiation. (a and b) Transverse retinal sections showing dual phosphorylated ERK 1/2inWT (a) and the lack of retinal activation of MAP kinase in yng mutant (b) eyes. (c) Example of focal intraocular injection. MAP kinase kinase (MEK) 1/2 inhibitors were injected intraocularly at 42 hpf, a time when ganglion cells have differentiated, but before photoreceptor differentiation. Ganglion cell and photoreceptor markers were assessed at 60 hpf. Phenol red is used as a visible marker to monitor injections. (d–f) Retinal sections of embryos injected with DMSO control (d) or MEK 1/2 inhibitors U0126 (e) and PD98059 (f) and then processed for a mixture of markers that indicate ganglion cell differentiation (zn8, arrows) and photoreceptor differentiation (zpr1, *). Note the absence of photoreceptor differentiation in embryos treated with either U0126 or PD98059. Ganglion cell markers were not affected, demonstrating the inhibitors did not alter retinal cell survival. (g) Quantitative comparison of photoreceptor differentiation between embryos with intraocular injections of DMSO, U0126, or PD98059. After statistical comparison, data were transformed to percent of control. *, P < 0.001, two-tailed Student′s t test; n = 8. |
|
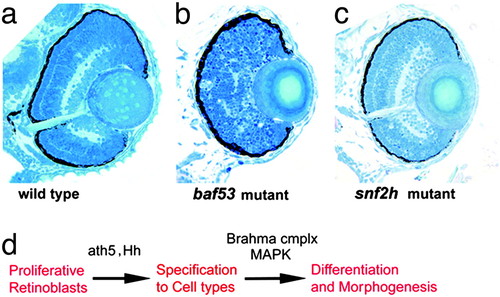
Analysis of additional chromatin remodeling mutants reveals specificity for function in retinal development. (a–c) Transverse sections through the central retina of WT (a), baf53 mutant hi550 (b), and snf2h mutant hi1072 (c). Baf53, which has been shown to complex with Brg1, has a retinal lamination phenotype similar to yng when mutated. snf2h, which encodes the ATPase for ISWI-type chromatin remodeling complexes, shows normal retinal differentiation. (d) Summary model: Ath5 and Hedgehog (Hh) are required for retinal cell type specification, whereas the Brahma complex is required for MAP kinase activation, which is necessary for retinal differentiation and morphogenesis. PHENOTYPE:
|

Unillustrated author statements PHENOTYPE:
|