- Title
-
olig2 is required for zebrafish primary motor neuron and oligodendrocyte development
- Authors
- Park, H.-C., Mehta, A., Richardson, J.S., and Appel, B.
- Source
- Full text @ Dev. Biol.
|
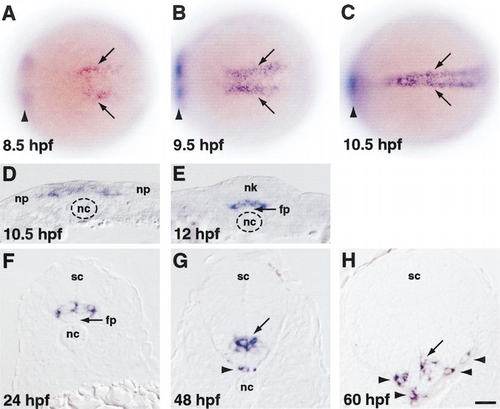
olig2 expression revealed by in situ RNA hybridization. (A–C) Dorsal views, anterior to the left, of whole embryos. Arrowheads indicate expression in prospective ventral diencephelon, which is out of the focal plane. (A) Prospective trunk spinal cord cells begin to express olig2 at midgastrula stage (arrows). (B) By late gastrula stage, olig2-expressing cells form two longitudinal stripes bordering the embryonic midline. (C) At neural plate stage, olig2-expressing cells form a single longitudinal domain at the midline. (D–H) Transverse sections, dorsal to the top, through trunk region. (D) Medial neural plate (np) cells, overlying notochord (nc), uniformly express olig2. (E) As neurulation proceeds, olig2-expressing cells occupy ventral neural keel (nk), overlying floor plate (fp). (F) After neurulation, cells within the ventral portion of the spinal cord (sc) express olig2. (G,H) Some olig2-expressing cells occupy basolateral positions, associated with the white matter (arrowheads). Scale bar equals 80 μm for (A–C) and 20 μm for (D–H). EXPRESSION / LABELING:
|
|
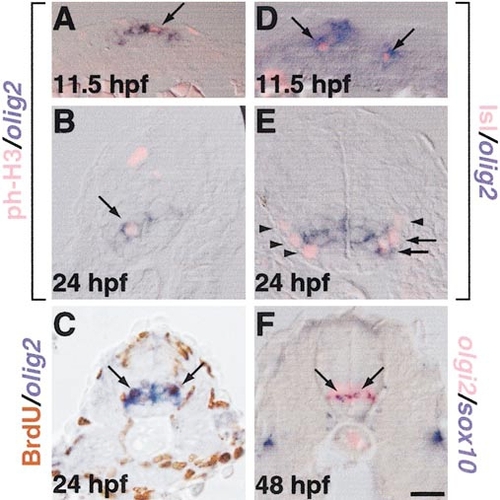
Proliferative neural cells, motor neurons, and oligodendrocytes express olig2. (A-F) Transverse sections through trunk regions of embryos probed for olig2 RNA expression by in situ hybridization. (A,B) M-phase cells, labeled by anti-phospho-histone H3 antibody (pink), and (C) S-phase cells, revealed by BrdU incorporation (brown) express olig2 (blue; arrows). (D) Primary motor neurons, labeled by anti-Is1 antibody (pink, arrows) express olig2 (blue, arrows). (E) At 24 hpf, some motor neurons express olig2 (arrows), whereas others do not (arrowheads). (F) At 48 hpf, olig2- positive cells (pink) include oligodendrocyte progenitors, marked by sox10 expression (blue, arrows). Scale bar, 20 μm. EXPRESSION / LABELING:
|
|
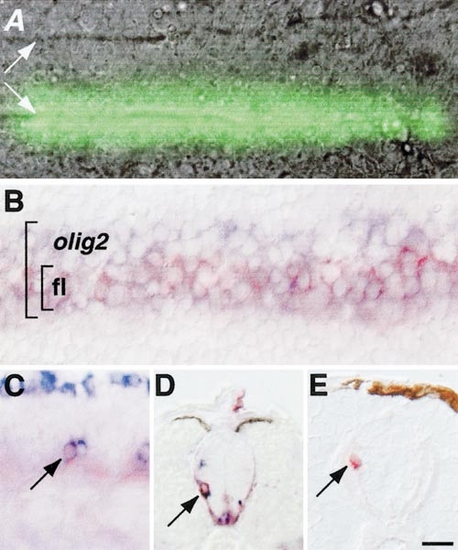
Fate mapping reveals that olig2-expressing neural plate cells give rise to primary motor neurons and oligodendrocytes (A) Dorsal view of living 10.5-hpf embryo in which caged fluorescein was photoactivated in a stripe of neuroectoderm overlying one edge of the notochord (notochord boundaries marked by white arrows). (B) Dorsal view of fixed 10.5-hpf embryo in which caged fluorescein had been photoactivated similarly to embryo in (A) and probed for olig2 RNA expression. Larger bracket indicates olig2-expressing cells, which are blue. Smaller bracket indicates cells in which caged fluorescein (fl) was photoactivated, revealed by red staining. Photoactivated cells are within the olig2 expression domain. (C) Side view of 19-hpf embryo probed for isl2 expression (blue) to reveal primary motor neurons and processed to reveal photoactivated fluorescein label (red). Photoactivated cell is a primary motor neuron (arrow). (D) Transverse section of 48-hpf embryo processed to reveal photoactivated fluorescein (red) and sox10 expression (blue). Arrow indicates double-labeled oligodendrocyte progenitor that originated in medial neural plate. (E) Transverse section of 72-hpf embryo processed to reveal photoactivated fluorescein label (red,arrow). Labeled cell occupies dorsal white matter, indicating that it is an oligodendrocyte, which originated in medial neural plate. Scale bar, 20 μm. EXPRESSION / LABELING:
|
|
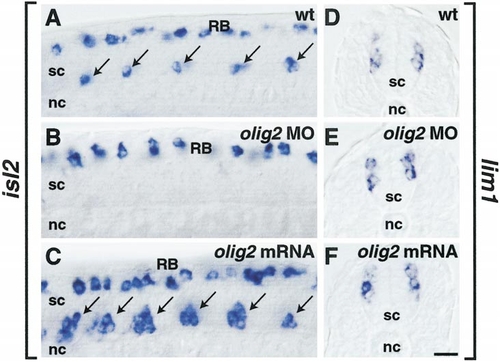
olig2 promotes primary motor neuron development. (A-C) Side views of 20-hpf embryos hybridized with isl2 probe. (A) Normal distribution of one to two primary motor neurons per ventral spinal cord (sc) hemisegment (arrows) and dorsal Rohon-Beard neurons (RB). (B) Embryo injected with olig2 MO lacked primary motor neurons but had Rohon-Beard neurons. (C) Embryo injected with olig2 mRNA. Excess primary motor neurons developed in ventral spinal cord (arrows), whereas Rohon-Beard neurons appeared normal. Apparent differences in Rohon-Beard cell number reflect normal variation and difference in depth of optical section. (D-F) Transverse sections of embryos hybridized with lim1 RNA probe. lim1-expressing cells appeared normal in embryos injected with olig2 MO (E) or olig2 mRNA (F). Scale bar, 20 μm for all panels. EXPRESSION / LABELING:
PHENOTYPE:
|
|
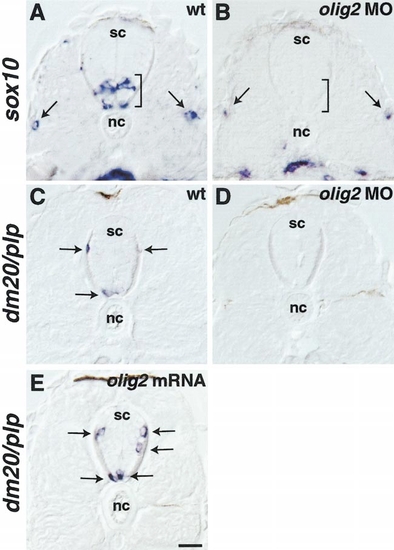
olig2 promotes oligodendrocyte development. (A, B) Transverse sections through trunk spinal cord (sc) of 48-hpf embryos hybridized with sox10 RNA probe. Brackets indicate ventral spinal cord and arrows point to sox10 expression in lateral line. Spinal cord expression of sox10 was absent from embryo injected with olig2 MO, whereas lateral line expression was normal (B). (C-E) Transverse sections of 72-hpf embryos hybridized with dm20/plp RNA probe. Arrows indicate dm20/plp-expressing oligodendrocytes. Embryos injected with olig2 MO did not express dm20/plp (D), whereas those injected with olig2 mRNA had a small but significant increase in the number of dm20/plp-expressing cells (E) (see Table 1). Scale bar, 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
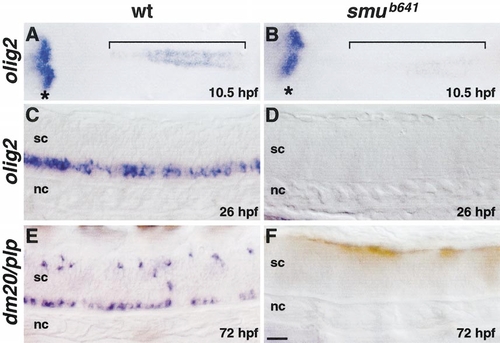
Hedgehog signaling is required for olig2 expression and oligodendrocyte development. (A, B) Dorsal views, anterior to the left, of flat-mounted embryos hybridized with olig2 probe. Brackets indicate olig2 expression in prospective spinal cord cells. smu mutant embryos express olig2 at a much lower level (B) compared with wild type (A). Asterisks mark olig2 expression in prospective ventral diencephelon, which appears normal in mutant embryos. (C, D) Side views of 26-hpf embryos, anterior to the left. Spinal cord cells of smu mutant embryos do not express olig2. (E, F) Side views of 72-hpf embryos, anterior to the left. Spinal cord oligodendrocytes, marked by dm20/plp expression, do not develop in smu mutant embryos. Scale bar, 40 μm (A and B) and 20 μm (C–F). EXPRESSION / LABELING:
PHENOTYPE:
|
|
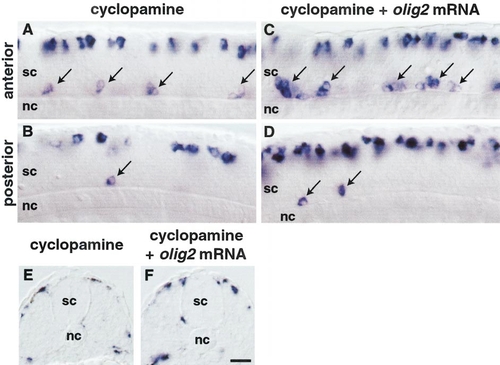
olig2 requires Hedgehog signaling to promote primary motor neuron and oligodendrocyte development. (A-D) Side views of 22-hpf embryos hybridized with isl2 probe. (A) Anterior and (B) posterior trunk spinal cord (sc) of embryo treated with cyclopamine. Slightly fewer isl2-positive primary motor neurons developed in anterior trunk relative to wild type (compare Fig. 8A with Fig. 5A), whereas few primary motor neurons developed in posterior trunk (B). (C) Anterior and (D) posterior trunk spinal cord of embryo injected with olig2 mRNA and treated with cyclopamine. Slightly more primary motor neurons developed in anterior trunk (C) compared with noninjected embryos (A), whereas primary motor neurons rarely developed in posterior trunk and tail (D), similar to noninjected embryos (B). (E, F) Transverse sections of 48-hpf embryos hybridized with sox10 probe. Spinal cord cells did not express sox10 in cyclopamine-treated embryos (E) or in olig2-injected embryos treated with cyclopamine (F). Scale bar, 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
Reprinted from Developmental Biology, 248(2), Park, H.-C., Mehta, A., Richardson, J.S., and Appel, B., olig2 is required for zebrafish primary motor neuron and oligodendrocyte development, 356-368, Copyright (2002) with permission from Elsevier. Full text @ Dev. Biol.