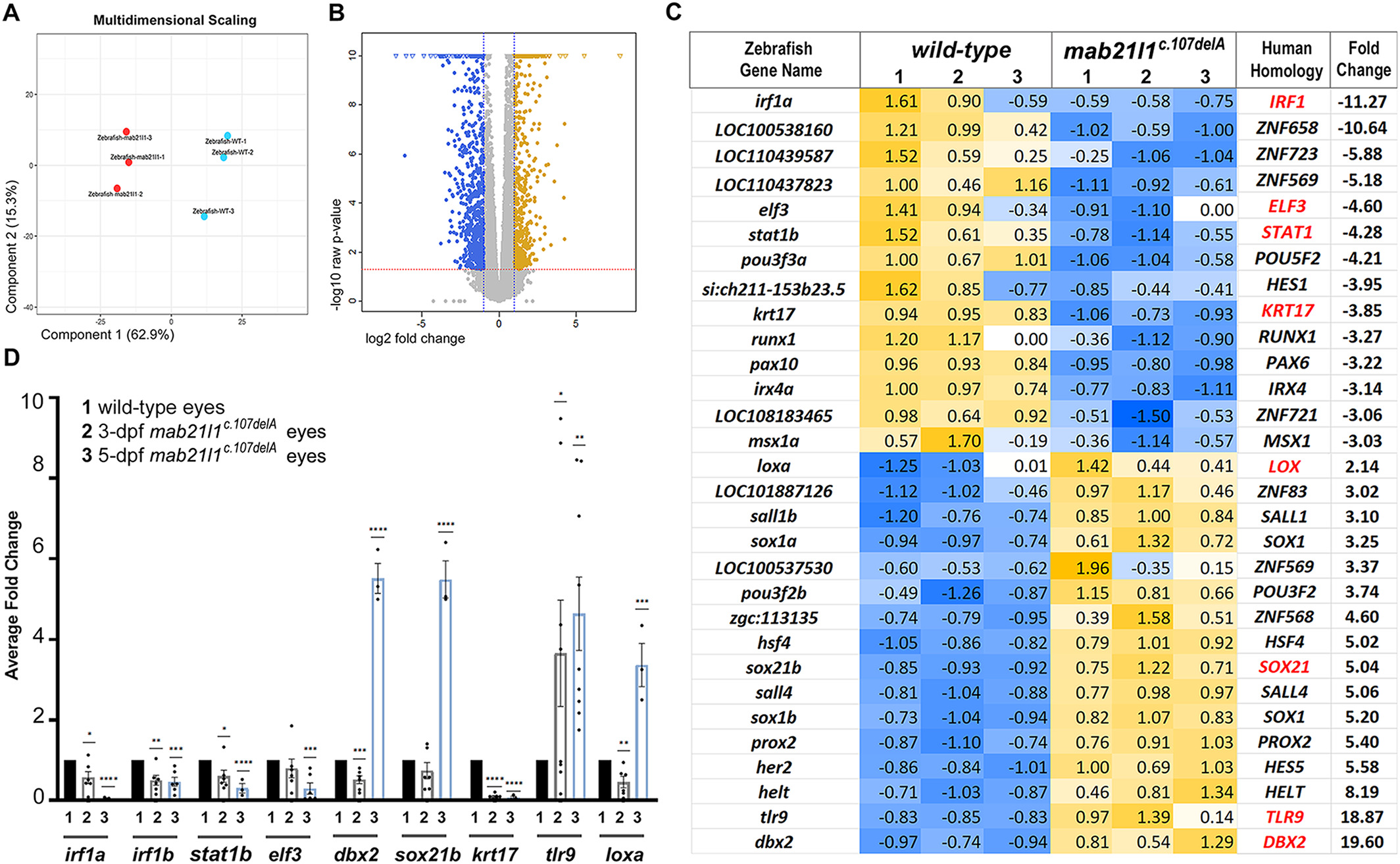
Fig. 6
Fig. 6 RNA seq data from wild type and mab21l1 mutant embryos. A, Multidimensional Scaling Analysis. Multidimensional scaling (MDS) plot using the gene expression values of each sample. B, Volcano Plot of Expression Level of the up-regulated (yellow) and down-regulated (blue) transcripts. Log2 fold change and P-value obtained from the comparison between two groups plotted as volcano plot. (X-axis: log2 Fold Change, Y-axis: -log10 P-value). The dashed red line shows the P-value cutoff (P = .01) with points above the line having P-value <.01 and points below the line having P-value >.01. The vertical dashed blue lines show twofold changes. C, Heatmap for differentially expressed transcription factors and related genes. Zebrafish gene name and homologous human gene name are indicated, along with fold change. Gene names in red indicate transcripts validated by qRT-PCR. All transcripts except for krt17, loxa and tlr9, encode transcription factors (please see text). D, qRT-PCR analysis of select transcript (irf1a, irf1b, stat1b, elf3, dbx2, sox21b, krt17, loxa, and tlr9) level in RNA extracted from whole eyes of 3- and 5-dpf wild-type and mab21l1c.107delA embryos. The expression level in mutant embryos was normalized to wild-type expression at the same time-point. Bar graphs for 1, 2, and 3 indicate wild-type, 3-dpf mab21l1c.107delA and 5-dpf mab21l1c.107delA, respectively. Statistical significance is indicated by asterisks; * P ≤ .05, ** P ≤ .01, *** P ≤ .001, ****P ≤ .0001; error bars indicate SEM